Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts
- PMID: 15317846
- PMCID: PMC524789
- DOI: 10.1091/mbc.e04-05-0405
Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts
Abstract
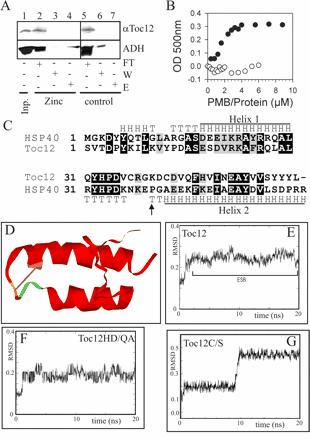
Translocation of proteins across membranes is essential for the biogenesis of each cell and is achieved by proteinaceous complexes. We analyzed the translocation complex of the intermembrane space from chloroplasts and identified a 12-kDa protein associated with the Toc machinery. Toc12 is an outer envelope protein exposing a soluble domain into the intermembrane space. Toc12 contains a J-domain and stimulates the ATPase activity of DnaK. The conformational stability and the ability to stimulate Hsp70 are dependent on a disulfide bridge within the loop region of the J-domain, suggesting a redox-regulated activation of the chaperone. Toc12 is associated with Toc64 and Tic22. Its J-domain recruits the Hsp70 of outer envelope membrane to the intermembrane space translocon and facilitates its interaction to the preprotein.
Figures




References
-
- Alexandrov, N.N., Nussinov, R., and Zimmer, R.M. (1995). Fast protein fold recognition via sequence to structure alignment and contact capacity potentials, Singapore: World Scientific Publishing, 53-72. - PubMed
-
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., and Hermans, J. (1981). Interaction models for water in relation to protein hydratation. In: Intermolecular Forces, ed. B. Pullman, Dordrecht: D. Reidel Publishing, 331-342.
-
- Berendsen, H.J.C., Postma, J.P.M., DiNola, A., and Haak, J.R. (1984). Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684-3690.
-
- Berendsen, H.J.C., van der Spoel, D., and van Drunen, R. (1995). GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Comm. 95, 43-56.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

