Transcription influences the types of deletion and expansion products in an orientation-dependent manner from GAC*GTC repeats
- PMID: 15317871
- PMCID: PMC516059
- DOI: 10.1093/nar/gkh787
Transcription influences the types of deletion and expansion products in an orientation-dependent manner from GAC*GTC repeats
Abstract
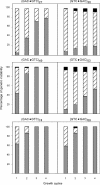
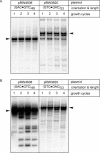
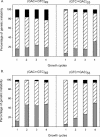
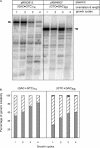
The genetic instability of (GAC*GTC)n (where n = 6-74) was investigated in an Escherichia coli-based plasmid system. Prior work implicated the instability of a (GAC*GTC)5 tract in the cartilage oligomeric matrix protein (COMP) gene to the 4, 6 or 7mers in the etiology of pseudoachondroplasia and multiple epiphyseal dysplasia. The effects of triplet repeat length and orientation were studied after multiple replication cycles in vivo. A transcribed plasmid containing (GAC*GTC)49 repeats led to large deletions (>3 repeats) after propagation in E.coli; however, if transcription was silenced by the LacI(Q) repressor, small expansions and deletions (<3 repeats) predominated the mutation spectra. In contrast, propagation of similar length but opposing orientation (GTC*GAC)53 containing plasmid led to small instabilities that were unaffected by the repression of transcription. Thus, by inhibiting transcription, the genetic instability of (GAC*GTC)49 repeats did not significantly differ from the opposing orientation, (GTC*GAC)53. We postulate that small instabilities of GAC*GTC repeats are achieved through replicative slippage, whereas large deletion events are found when GAC*GTC repeats are transcribed. Herein, we report the first genetic study on GAC*GTC repeat instability describing two types of mutational patterns that can be partitioned by transcription modulation. Along with prior biophysical data, these results lay the initial groundwork for understanding the genetic processes responsible for triplet repeat mutations in the COMP gene.
Figures


References
-
- Wells R.D. and Warren,S.T. (eds) (1998) Genetic Instabilities and Hereditary Neurological Diseases. Academic Press, San Diego, CA.
-
- Cummings C.J. and Zoghbi,H.Y. (2000) Trinucleotide repeats: mechanisms and pathophysiology. Annu. Rev. Genomics Hum. Genet., 1, 281–328. - PubMed
-
- Delot E., King,L.M., Briggs,M.D., Wilcox,W.R. and Cohn,D.H. (1999) Trinucleotide expansion mutations in the cartilage oligomeric matrix protein (COMP) gene. Hum. Mol. Genet., 8, 123–128. - PubMed
-
- Ohshima K., Kang,S. and Wells,R.D. (1996) CTG triplet repeats from human hereditary diseases are dominant genetic expansion products in Escherichia coli. J. Biol. Chem., 271, 1853–1856. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

