The contribution of metal ions to the structural stability of the large ribosomal subunit
- PMID: 15317974
- PMCID: PMC1370624
- DOI: 10.1261/rna.7390804
The contribution of metal ions to the structural stability of the large ribosomal subunit
Abstract
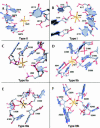
Both monovalent cations and magnesium ions are well known to be essential for the folding and stability of large RNA molecules that form complex and compact structures. In the atomic structure of the large ribosomal subunit from Haloarcula marismortui, we have identified 116 magnesium ions and 88 monovalent cations bound principally to rRNA. Although the rRNA structures to which these metal ions bind are highly idiosyncratic, a few common principles have emerged from the identities of the specific functional groups that coordinate them. The nonbridging oxygen of a phosphate group is the most common inner shell ligand of Mg++, and Mg++ ions having one or two such inner shell ligands are very common. Nonbridging phosphate oxygens and the heteroatoms of nucleotide bases are common outer shell ligands for Mg++ ions. Monovalent cations usually interact with nucleotide bases and protein groups, although some interactions with nonbridging phosphate oxygens are found. The most common monovalent cation binding site is the major groove side of G-U wobble pairs. Both divalent and monovalent cations stabilize the tertiary structure of 23S rRNA by mediating interactions between its structural domains. Bound metal ions are particularly abundant in the region surrounding the peptidyl transferase center, where stabilizing cationic tails of ribosomal proteins are notably absent. This may point to the importance of metal ions for the stabilization of specific RNA structures in the evolutionary period prior to the appearance of proteins, and hence many of these metal ion binding sites may be conserved across all phylogenetic kingdoms.
Figures







References
-
- Auffinger, P., Bielecki, L., and Westhof, E. 2004. Anion binding to nucleic acids. Structure 12: 379–388. - PubMed
-
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. - PubMed
-
- Basu, S., Rambo, R.P., Strauss-Soukup, J., Cate, J.H., Ferre-D’Amare, A.R., Strobel, S.A., and Doudna, J.A. 1998. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat. Struct. Biol. 5: 986–992. - PubMed
-
- Batey, R.T. and Doudna, J.A. 2002. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry 41: 11703–11710. - PubMed
-
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
