Tissue-specific modification of gld-2 mRNA in C. elegans: likely C-to-U editing
- PMID: 15317977
- PMCID: PMC1370630
- DOI: 10.1261/rna.7570804
Tissue-specific modification of gld-2 mRNA in C. elegans: likely C-to-U editing
Abstract
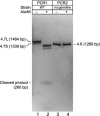
Seventeen years after the discovery of tissue-specific apoB mRNA editing, only three nucleus-encoded mRNAs have been shown to undergo C-to-U editing. All three mRNAs occur in mammals. apoB mRNA editing is tissue-specific and occurs normally, whereas NF1 and NAT1 mRNA editing is found largely in tumors. Here we report the first example of C-to-U RNA editing in Caenorhabditis elegans. The gld-2 gene encodes an atypical poly(A) polymerase that governs the mitosis/meiosis decision in the germ line as well as progression through meiosis and early embryogenesis. At least two of its alternatively spliced transcripts are germline-specific. We find that most and perhaps all germline-specific transcripts generated by the gld-2 gene undergo C-to-U editing, but that somatic transcripts show no detectable editing. The gld-2 C-to-U editing event changes the codon from CCG to CUG, which is predicted to cause a proline to leucine substitution in the protein sequence. Our findings suggest the presence of a sequence- and tissue-specific cytidine deaminase acting on RNA, or CDAR. This CDAR modifies a specific base in gld-2 mRNA, and acts only in the germline.
Figures

Similar articles
-
A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans.Nature. 2002 Sep 19;419(6904):312-6. doi: 10.1038/nature01039. Nature. 2002. PMID: 12239571
-
Gene regulation: reviving the message.Nature. 2002 Sep 19;419(6904):267-8. doi: 10.1038/419267a. Nature. 2002. PMID: 12239557 No abstract available.
-
GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans.Genetics. 2004 Sep;168(1):147-60. doi: 10.1534/genetics.104.029264. Genetics. 2004. PMID: 15454534 Free PMC article.
-
The regulatory network controlling the proliferation-meiotic entry decision in the Caenorhabditis elegans germ line.Curr Top Dev Biol. 2006;76:185-215. doi: 10.1016/S0070-2153(06)76006-9. Curr Top Dev Biol. 2006. PMID: 17118267 Review.
-
C. elegans star proteins, GLD-1 and ASD-2, regulate specific RNA targets to control development.Adv Exp Med Biol. 2010;693:106-22. doi: 10.1007/978-1-4419-7005-3_8. Adv Exp Med Biol. 2010. PMID: 21189689 Review.
Cited by
-
mRNA Editing, Processing and Quality Control in Caenorhabditis elegans.Genetics. 2020 Jul;215(3):531-568. doi: 10.1534/genetics.119.301807. Genetics. 2020. PMID: 32632025 Free PMC article. Review.
-
The C. elegans embryonic transcriptome with tissue, time, and alternative splicing resolution.Genome Res. 2019 Jun;29(6):1036-1045. doi: 10.1101/gr.243394.118. Epub 2019 May 23. Genome Res. 2019. PMID: 31123079 Free PMC article.
-
Nucleotide levels regulate germline proliferation through modulating GLP-1/Notch signaling in C. elegans.Genes Dev. 2016 Feb 1;30(3):307-20. doi: 10.1101/gad.275107.115. Genes Dev. 2016. PMID: 26833730 Free PMC article.
-
Hippocampal Characteristics and Invariant Sequence Elements Distribution of GLRA2 and GLRA3 C-to-U Editing.Mol Syndromol. 2017 Mar;8(2):85-92. doi: 10.1159/000453300. Epub 2016 Dec 16. Mol Syndromol. 2017. PMID: 28611548 Free PMC article.
-
Profiling the RNA editomes of wild-type C. elegans and ADAR mutants.Genome Res. 2015 Jan;25(1):66-75. doi: 10.1101/gr.176107.114. Epub 2014 Nov 4. Genome Res. 2015. PMID: 25373143 Free PMC article.
References
-
- Bass, B.L. 1993. RNA editing: New uses for old players in the RNA world. In The RNA world (eds. R. Gesteland and J. Atkins), pp. 383–418. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Benne, R., Van den Burg, J., Brakenhoff, J.P., Sloof, P., Van Boom, J.H., and Tromp, M.C. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46: 819–826. - PubMed
-
- Blanc, V. and Davidson, N.O. 2003. C-to-U RNA editing: Mechanisms leading to genetic diversity. J. Biol. Chem. 278: 1395–1398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
