Transcriptomic changes in human breast cancer progression as determined by serial analysis of gene expression
- PMID: 15318932
- PMCID: PMC549167
- DOI: 10.1186/bcr899
Transcriptomic changes in human breast cancer progression as determined by serial analysis of gene expression
Erratum in
- Breast Cancer Res. 2005;7(1):32
Abstract
Introduction: Genomic and transcriptomic alterations affecting key cellular processes such us cell proliferation, differentiation and genomic stability are considered crucial for the development and progression of cancer. Most invasive breast carcinomas are known to derive from precursor in situ lesions. It is proposed that major global expression abnormalities occur in the transition from normal to premalignant stages and further progression to invasive stages. Serial analysis of gene expression (SAGE) was employed to generate a comprehensive global gene expression profile of the major changes occurring during breast cancer malignant evolution.
Methods: In the present study we combined various normal and tumor SAGE libraries available in the public domain with sets of breast cancer SAGE libraries recently generated and sequenced in our laboratory. A recently developed modified t test was used to detect the genes differentially expressed.
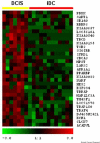
Results: We accumulated a total of approximately 1.7 million breast tissue-specific SAGE tags and monitored the behavior of more than 25,157 genes during early breast carcinogenesis. We detected 52 transcripts commonly deregulated across the board when comparing normal tissue with ductal carcinoma in situ, and 149 transcripts when comparing ductal carcinoma in situ with invasive ductal carcinoma (P < 0.01).
Conclusion: A major novelty of our study was the use of a statistical method that correctly accounts for the intra-SAGE and inter-SAGE library sources of variation. The most useful result of applying this modified t statistics beta binomial test is the identification of genes and gene families commonly deregulated across samples within each specific stage in the transition from normal to preinvasive and invasive stages of breast cancer development. Most of the gene expression abnormalities detected at the in situ stage were related to specific genes in charge of regulating the proper homeostasis between cell death and cell proliferation. The comparison of in situ lesions with fully invasive lesions, a much more heterogeneous group, clearly identified as the most importantly deregulated group of transcripts those encoding for various families of proteins in charge of extracellular matrix remodeling, invasion and cell motility functions.
Figures



Similar articles
-
Evidence that molecular changes in cells occur before morphological alterations during the progression of breast ductal carcinoma.Breast Cancer Res. 2008;10(5):R87. doi: 10.1186/bcr2157. Epub 2008 Oct 17. Breast Cancer Res. 2008. PMID: 18928525 Free PMC article.
-
Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer.Cancer Res. 2012 Sep 1;72(17):4574-86. doi: 10.1158/0008-5472.CAN-12-0636. Epub 2012 Jul 2. Cancer Res. 2012. PMID: 22751464 Free PMC article.
-
HIN-1, a putative cytokine highly expressed in normal but not cancerous mammary epithelial cells.Proc Natl Acad Sci U S A. 2001 Aug 14;98(17):9796-801. doi: 10.1073/pnas.171138398. Epub 2001 Jul 31. Proc Natl Acad Sci U S A. 2001. PMID: 11481438 Free PMC article.
-
Molecular markers for the diagnosis and management of ductal carcinoma in situ.J Natl Cancer Inst Monogr. 2010;2010(41):210-3. doi: 10.1093/jncimonographs/lgq019. J Natl Cancer Inst Monogr. 2010. PMID: 20956832 Free PMC article. Review.
-
Carcinoma in situ of the female breast. A clinico-pathological, immunohistological, and DNA ploidy study.APMIS Suppl. 2003;(108):1-67. APMIS Suppl. 2003. PMID: 12874968 Review.
Cited by
-
The feedback loop of LITAF and BCL6 is involved in regulating apoptosis in B cell non-Hodgkin's-lymphoma.Oncotarget. 2016 Nov 22;7(47):77444-77456. doi: 10.18632/oncotarget.12680. Oncotarget. 2016. PMID: 27764808 Free PMC article.
-
Role of heparan sulfatases in ovarian and breast cancer.Am J Cancer Res. 2013;3(1):34-45. Epub 2013 Jan 18. Am J Cancer Res. 2013. PMID: 23359864 Free PMC article.
-
Evaluating Serum Markers for Hormone Receptor-Negative Breast Cancer.PLoS One. 2015 Nov 13;10(11):e0142911. doi: 10.1371/journal.pone.0142911. eCollection 2015. PLoS One. 2015. PMID: 26565788 Free PMC article.
-
Chemical carcinogen-induced rat mammary carcinogenesis is a potential model of p21-activated kinase positive female breast cancer.Physiol Genomics. 2021 Feb 1;53(2):61-68. doi: 10.1152/physiolgenomics.00112.2020. Epub 2020 Dec 21. Physiol Genomics. 2021. PMID: 33346690 Free PMC article.
-
Stromal genes add prognostic information to proliferation and histoclinical markers: a basis for the next generation of breast cancer gene signatures.PLoS One. 2012;7(6):e37646. doi: 10.1371/journal.pone.0037646. Epub 2012 Jun 18. PLoS One. 2012. PMID: 22719844 Free PMC article.
References
-
- Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics. CA Cancer J Clin. 2000;50:7–33. - PubMed
-
- Berardo MD, Allred DC, O'Connell P. Breast cancer. In: Jameson JL, editor. In Principles of Molecular Medicine. Totowa, NJ: Human Press; 1998. pp. 625–632.
-
- Ponten J, Holmberg L, Trichopoulos D, Kallioniemi O, Kvale G, Wallgren A. Biology and natural history of breast cancer. Int J Cancer. 1990;5:5–21. - PubMed
-
- Charpentier A, Aldaz CM. The molecular basis of breast carcinogenesis. In: Coleman WB, Tsongalis GJ, editor. In The Molecular Basis of Human Cancer. Totowa, NJ: Human Press; 2002. pp. 347–363.
-
- Troup S, Njue C, Kliewer EV, Parisien M, Roskelley C, Chakravarti S, Roughley PJ, Murphy LC, Watson PH. Reduced expression of the small leucine-rich proteoglycans, Lumican, and Decorin is associated with poor outcome in node-negative breast cancer. Clin Cancer Res. 2003;9:207–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

