A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice
- PMID: 15319478
- PMCID: PMC520948
- DOI: 10.1105/tpc.104.022715
A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice
Abstract
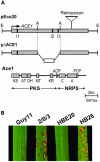
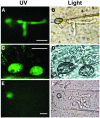
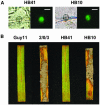
Isolates of the rice blast fungus Magnaporthe grisea that carry the gene encoding Avirulence Conferring Enzyme1 (ACE1) are specifically recognized by rice (Oryza sativa) cultivars carrying the resistance gene Pi33. This recognition enables resistant plants to activate a defense response. ACE1 was isolated by map-based cloning and encodes a putative hybrid between a polyketide synthase and a nonribosomal peptide synthetase, enzymes involved in microbial secondary metabolism. ACE1 is expressed exclusively during fungal penetration of host leaves, the time point at which plant defense reactions are triggered. Ace1 appears to be localized in the cytoplasm of the appressorium. Mutation of the putative catalytic site of the beta-ketoacyl synthase domain of Ace1 abolishes recognition of the fungus by resistant rice. This suggests that Ace1 biosynthetic activity is required for avirulence. Our results are consistent with the hypothesis that the fungal signal recognized by resistant rice plants is the secondary metabolite whose synthesis depends on Ace1.
Figures


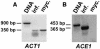
References
-
- Abe, Y., Suzuki, T., Ono, C., Iwamoto, K., Hosobuchi, M., and Yoshikawa, H. (2002). Molecular cloning and characterization of an ML-236B (compactin) biosynthetic gene cluster in Penicillium citrinum. Mol. Genet. Genomics 267, 636–646. - PubMed
-
- Aparicio, J.F., Molnar, I., Schwecke, T., Konig, A., Haydock, S.F., Khaw, L.E., Staunton, J., and Leadlay, P.F. (1996). Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: Analysis of the enzymatic domains in the modular polyketide synthase. Gene 169, 9–16. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

