From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants
- PMID: 15319480
- PMCID: PMC520932
- DOI: 10.1105/tpc.104.023135
From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants
Abstract
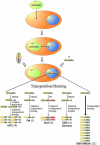
All members of the AP2/ERF family of plant transcription regulators contain at least one copy of a DNA binding domain called the AP2 domain. The AP2 domain has been considered plant specific. Here, we show that homologs are present in the cyanobacterium Trichodesmium erythraeum, the ciliate Tetrahymena thermophila, and the viruses Enterobacteria phage Rb49 and Bacteriophage Felix 01. We demonstrate that the T. erythraeum AP2 domain selectively binds stretches of poly(dG)/poly(dC) showing functional conservation with plant AP2/ERF proteins. The newly discovered nonplant proteins bearing an AP2 domain are predicted to be HNH endonucleases. Sequence conservation extends outside the AP2 domain to include part of the endonuclease domain for the T. erythraeum protein and some plant AP2/ERF proteins. Our phylogenetic analysis of the broader family of AP2 domains supports the possibility of lateral gene transfer. We hypothesize that a horizontal transfer of an HNH-AP2 endonuclease from bacteria or viruses into plants may have led to the origin of the AP2/ERF family of transcription factors via transposition and homing processes.
Figures





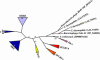

References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Bailey, T.L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. (Menlo Park, CA: AAAI Press), pp. 28–36. - PubMed
-
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

