The Arabidopsis transthyretin-like protein is a potential substrate of BRASSINOSTEROID-INSENSITIVE 1
- PMID: 15319482
- PMCID: PMC520942
- DOI: 10.1105/tpc.104.023903
The Arabidopsis transthyretin-like protein is a potential substrate of BRASSINOSTEROID-INSENSITIVE 1
Abstract
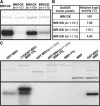
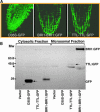
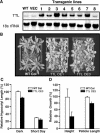
BRASSINOSTEROID-INSENSITIVE 1 (BRI1) is a Leu-rich-repeat (LRR) receptor kinase that functions as a critical component of a transmembrane brassinosteroid (BR) receptor. It is believed that BRI1 becomes activated through heterodimerization with BAK1, a similar LRR receptor kinase, in response to BR signal. A yeast two-hybrid screen using the kinase domain of BRI1 identified an Arabidopsis thaliana Transthyretin-Like protein (TTL) as a potential BRI1 substrate. TTL interacts with BRI1 in a kinase-dependent manner in yeast and is phosphorylated by BRI1 in vitro. TTL displays a similar expression pattern with BRI1 and is associated with the plasma membrane. Overexpression of the TTL gene results in a phenotype that was observed in weak bri1 mutants and null bak1 mutants. By contrast, two T-DNA insertional mutations in the TTL gene promote plant growth and enhance BR sensitivity. We hypothesized that TTL might directly regulate certain biochemical activities near the plasma membrane to control plant growth.
Figures




References
-
- Aranda, A., and Pascual, A. (2001). Nuclear hormone receptors and gene expression. Physiol. Rev. 81, 1269–1304. - PubMed
-
- Benson, M.D., and Uemichi, T. (1996). Transthyretin amyloidosis amyloid. Int. J. Exp. Clin. Invest. 3, 44–56.
-
- Caño-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-García, S., Cheng, J.-C., Nam, K.H., Li, J., and Chory, J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development, in press. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

