Activation of PPARgamma is required for curcumin to induce apoptosis and to inhibit the expression of extracellular matrix genes in hepatic stellate cells in vitro
- PMID: 15320868
- PMCID: PMC1134098
- DOI: 10.1042/BJ20040928
Activation of PPARgamma is required for curcumin to induce apoptosis and to inhibit the expression of extracellular matrix genes in hepatic stellate cells in vitro
Abstract
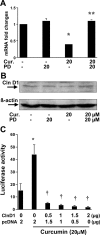
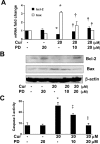
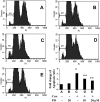
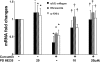
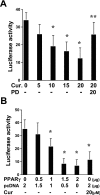
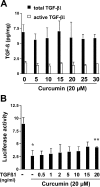
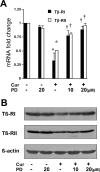
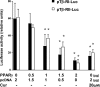
During liver fibrogenesis, quiescent HSC (hepatic stellate cells) become active, a transformation that is associated with enhanced cell proliferation and overproduction of ECM (extracellular matrix). Inhibition of cell proliferation and induction of apoptosis are potential strategies to block the activation of HSC for the prevention and treatment of liver fibrosis. Levels of PPARgamma (peroxisome proliferator-activated receptor gamma) are dramatically diminished in parallel with HSC activation. Stimulation of PPARgamma by its agonists inhibits HSC activation in vitro and in vivo. We demonstrated recently that curcumin, the yellow pigment in curry, inhibited HSC activation in vitro, reducing cell proliferation, inducing apoptosis and inhibiting ECM gene expression. Further studies indicated that curcumin induced the gene expression of PPARgamma and stimulated its activity in activated HSC in vitro, which was required for curcumin to inhibit HSC proliferation. The aims of the present study were to evaluate the roles of PPARgamma activation in the induction of apoptosis and suppression of ECM gene expression by curcumin in activated HSC, and to elucidate the underlying mechanisms. Our results demonstrated that blocking PPARgamma activation abrogated the effects of curcumin on the induction of apoptosis and inhibition of the expression of ECM genes in activated HSC in vitro. Further experiments demonstrated that curcumin suppressed the gene expression of TGF-beta (transforming growth factor-beta) receptors and interrupted the TGF-beta signalling pathway in activated HSC, which was mediated by PPARgamma activation. Taken together, our results demonstrate that curcumin stimulated PPARgamma activity in activated HSC in vitro, which was required for curcumin to reduce cell proliferation, induce apoptosis and suppress ECM gene expression. These results provide novel insight into the mechanisms responsible for the inhibition of HSC activation by curcumin. The characteristics of curcumin, which has no adverse health effects, make it a potential candidate for prevention and treatment of hepatic fibrosis.
Figures

Similar articles
-
Disruption of transforming growth factor-beta signaling by curcumin induces gene expression of peroxisome proliferator-activated receptor-gamma in rat hepatic stellate cells.Am J Physiol Gastrointest Liver Physiol. 2007 Jan;292(1):G113-23. doi: 10.1152/ajpgi.00200.2006. Epub 2006 Sep 7. Am J Physiol Gastrointest Liver Physiol. 2007. PMID: 16959952
-
Activation of PPARγ/P53 signaling is required for curcumin to induce hepatic stellate cell senescence.Cell Death Dis. 2016 Apr 14;7(4):e2189. doi: 10.1038/cddis.2016.92. Cell Death Dis. 2016. PMID: 27077805 Free PMC article.
-
Activation of peroxisome proliferator-activated receptor-gamma contributes to the inhibitory effects of curcumin on rat hepatic stellate cell growth.Am J Physiol Gastrointest Liver Physiol. 2003 Jul;285(1):G20-30. doi: 10.1152/ajpgi.00474.2002. Epub 2003 Mar 26. Am J Physiol Gastrointest Liver Physiol. 2003. PMID: 12660143
-
Wnt signaling in liver fibrosis: progress, challenges and potential directions.Biochimie. 2013 Dec;95(12):2326-35. doi: 10.1016/j.biochi.2013.09.003. Epub 2013 Sep 13. Biochimie. 2013. PMID: 24036368 Review.
-
Organ fibrosis inhibited by blocking transforming growth factor-β signaling via peroxisome proliferator-activated receptor γ agonists.Hepatobiliary Pancreat Dis Int. 2012 Oct;11(5):467-78. doi: 10.1016/s1499-3872(12)60210-0. Hepatobiliary Pancreat Dis Int. 2012. PMID: 23060391 Review.
Cited by
-
Curcumin inhibits gene expression of receptor for advanced glycation end-products (RAGE) in hepatic stellate cells in vitro by elevating PPARγ activity and attenuating oxidative stress.Br J Pharmacol. 2012 Aug;166(8):2212-27. doi: 10.1111/j.1476-5381.2012.01910.x. Br J Pharmacol. 2012. PMID: 22352842 Free PMC article.
-
Curcumin promotes cholesterol efflux from adipocytes related to PPARgamma-LXRalpha-ABCA1 passway.Mol Cell Biochem. 2011 Dec;358(1-2):281-5. doi: 10.1007/s11010-011-0978-z. Epub 2011 Jul 12. Mol Cell Biochem. 2011. PMID: 21748336
-
Activated PPARgamma Targets Surface and Intracellular Signals That Inhibit the Proliferation of Lung Carcinoma Cells.PPAR Res. 2008;2008:254108. doi: 10.1155/2008/254108. PPAR Res. 2008. PMID: 18704200 Free PMC article.
-
De novo synthesis of glutathione is a prerequisite for curcumin to inhibit hepatic stellate cell (HSC) activation.Free Radic Biol Med. 2007 Aug 1;43(3):444-53. doi: 10.1016/j.freeradbiomed.2007.04.016. Epub 2007 Apr 29. Free Radic Biol Med. 2007. PMID: 17602960 Free PMC article.
-
A Comprehensive Review of Natural Products against Liver Fibrosis: Flavonoids, Quinones, Lignans, Phenols, and Acids.Evid Based Complement Alternat Med. 2020 Oct 5;2020:7171498. doi: 10.1155/2020/7171498. eCollection 2020. Evid Based Complement Alternat Med. 2020. PMID: 33082829 Free PMC article. Review.
References
-
- Friedman S. L. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J. Biol. Chem. 2000;275:2247–2250. - PubMed
-
- Schuppan D., Atkinson J., Ruehl M., Riecken E. O. Alcohol and liver fibrosis – pathobiochemistry and treatment. Z. Gastroenterol. 1995;33:546–550. - PubMed
-
- Dooley S., Delvoux B., Lahme B., Mangasser-Stephan K., Gressner A. M. Modulation of transforming growth factor beta response and signaling during transdifferentiation of rat hepatic stellate cells to myofibroblasts. Hepatology. 2000;31:1094–1106. - PubMed
-
- Friedman S. L., Yamasaki G., Wong L. Modulation of transforming growth factor beta receptors of rat lipocytes during the hepatic wound healing response. Enhanced binding and reduced gene expression accompany cellular activation in culture and in vivo. J. Biol. Chem. 1994;269:10551–10558. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

