Contribution of the two conserved tryptophan residues to the catalytic and structural properties of Proteus mirabilis glutathione S-transferase B1-1
- PMID: 15320869
- PMCID: PMC1134671
- DOI: 10.1042/BJ20040890
Contribution of the two conserved tryptophan residues to the catalytic and structural properties of Proteus mirabilis glutathione S-transferase B1-1
Abstract
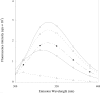
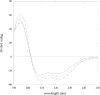
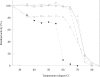
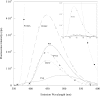
PmGSTB1-1 (Proteus mirabilis glutathione S-transferase B1-1) has two tryptophan residues at positions 97 and 164 in each monomer. Structural data for this bacterial enzyme indicated that Trp97 is positioned in the helix a4, whereas Trp164 is located at the bottom of the helix a6 in the xenobiotic-binding site. To elucidate the role of the two tryptophan residues they were replaced by site-directed mutagenesis. Trp97 and Trp164 were mutated to either phenylalanine or alanine. A double mutant was also constructed. The effects of the replacement on the activity, structural properties and antibiotic-binding capacity of the enzymes were examined. On the basis of the results obtained, Trp97 does not seem to be involved in the enzyme active site and structural stabilization. In contrast, different results were achieved for Trp164 mutants. Conservative substitution of the Trp164 with phenylalanine enhanced enzyme activity 10-fold, whereas replacement with alanine enhanced enzyme activity 17-fold. Moreover, the catalytic efficiency for both GSH and 1-chloro-2,4-dinitrobenzene substrates improved. In particular, the catalytic efficiency for 1-chloro-2,4-dinitrobenzene improved for both W164F (Trp164-->Phe) and W164A by factors of 7- and 22-fold respectively. These results are supported by molecular graphic analysis. In fact, W164A presented a more extensive substrate-binding pocket that could allow the substrates to be better accommodated. Furthermore, both Trp164 mutants were significantly more thermolabile than wild-type, suggesting that the substitution of this residue affects the overall stability of the enzyme. Taken together, these results indicate that Trp164 is an important residue of PmGSTB1-1 in the catalytic process as well as for protein stability.
Figures


Similar articles
-
Evolutionarily conserved structural motifs in bacterial GST (glutathione S-transferase) are involved in protein folding and stability.Biochem J. 2006 Feb 15;394(Pt 1):11-7. doi: 10.1042/BJ20051367. Biochem J. 2006. PMID: 16248855 Free PMC article.
-
Evaluation of the role of two conserved active-site residues in beta class glutathione S-transferases.Biochem J. 2000 Oct 15;351 Pt 2(Pt 2):341-6. Biochem J. 2000. PMID: 11023819 Free PMC article.
-
Characterization of the hydrophobic substrate-binding site of the bacterial beta class glutathione transferase from Proteus mirabilis.Protein Eng Des Sel. 2010 Sep;23(9):743-50. doi: 10.1093/protein/gzq048. Epub 2010 Jul 27. Protein Eng Des Sel. 2010. PMID: 20663851
-
Glutamic acid-65 is an essential residue for catalysis in Proteus mirabilis glutathione S-transferase B1-1.Biochem J. 2002 Apr 1;363(Pt 1):189-93. doi: 10.1042/0264-6021:3630189. Biochem J. 2002. PMID: 11903062 Free PMC article.
-
Production of phenylpyruvic acid by engineered L-amino acid deaminase from Proteus mirabilis.Biotechnol Lett. 2022 Jun;44(5-6):635-642. doi: 10.1007/s10529-022-03245-y. Epub 2022 Apr 16. Biotechnol Lett. 2022. PMID: 35429303 Review.
Cited by
-
Identification of an elongation factor 1Bγ protein with glutathione transferase activity in both yeast and mycelial morphologies from human pathogenic Blastoschizomyces capitatus.Folia Microbiol (Praha). 2014 Mar;59(2):107-13. doi: 10.1007/s12223-013-0273-3. Epub 2013 Aug 3. Folia Microbiol (Praha). 2014. PMID: 23913100
-
Evolutionarily conserved structural motifs in bacterial GST (glutathione S-transferase) are involved in protein folding and stability.Biochem J. 2006 Feb 15;394(Pt 1):11-7. doi: 10.1042/BJ20051367. Biochem J. 2006. PMID: 16248855 Free PMC article.
References
-
- Hayes J. D., Pulford D. J. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. - PubMed
-
- Hayes J. D., McLellan L. I. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radical Res. 1999;31:273–300. - PubMed
-
- Armstrong R. N. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 1997;10:2–18. - PubMed
-
- Mannervik B., Danielson U. H. Glutathione transferases: structure and catalytic activity. Crit. Rev. Biochem. Mol. Biol. 1988;23:283–337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

