Conformational stability and thermodynamic characterization of the lipoic acid bearing domain of human mitochondrial branched chain alpha-ketoacid dehydrogenase
- PMID: 15322287
- PMCID: PMC2280005
- DOI: 10.1110/ps.04783104
Conformational stability and thermodynamic characterization of the lipoic acid bearing domain of human mitochondrial branched chain alpha-ketoacid dehydrogenase
Abstract
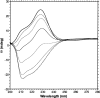
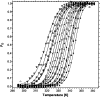
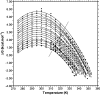
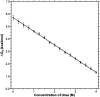
The lipoic acid bearing domain (hbLBD) of human mitochondrial branched chain alpha-ketoacid dehydrogenase (BCKD) plays important role of substrate channeling in oxidative decarboxylation of the branched chain alpha-ketoacids. Recently hbLBD has been found to follow two-step folding mechanism without detectable presence of stable or kinetic intermediates. The present study describes the conformational stability underlying the folding of this small beta-barrel domain. Thermal denaturation in presence of urea and isothermal urea denaturation titrations are used to evaluate various thermodynamic parameters defining the equilibrium unfolding. The linear extrapolation model successfully describes the two-step; native state <-->denatured state unfolding transition of hbLBD. The average temperature of maximum stability of hbLBD is estimated as 295.6 +/- 0.9 K. Cold denaturation of hbLBD is also predicted and discussed.
Figures



Similar articles
-
Folding of the protein domain hbSBD.Biophys J. 2005 Nov;89(5):3353-61. doi: 10.1529/biophysj.105.065151. Epub 2005 Aug 26. Biophys J. 2005. PMID: 16126825 Free PMC article.
-
Folding kinetics of the lipoic acid-bearing domain of human mitochondrial branched chain alpha-ketoacid dehydrogenase complex.FEBS Lett. 2002 Oct 23;530(1-3):133-8. doi: 10.1016/s0014-5793(02)03444-0. FEBS Lett. 2002. PMID: 12387880
-
Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex.J Biol Chem. 2002 May 3;277(18):15865-73. doi: 10.1074/jbc.M110952200. Epub 2002 Feb 11. J Biol Chem. 2002. PMID: 11839747
-
Plasmodium falciparum possesses organelle-specific alpha-keto acid dehydrogenase complexes and lipoylation pathways.Biochem Soc Trans. 2005 Nov;33(Pt 5):977-80. doi: 10.1042/BST20050977. Biochem Soc Trans. 2005. PMID: 16246025 Review.
-
Lipoic acid metabolism and mitochondrial redox regulation.J Biol Chem. 2018 May 18;293(20):7522-7530. doi: 10.1074/jbc.TM117.000259. Epub 2017 Nov 30. J Biol Chem. 2018. PMID: 29191830 Free PMC article. Review.
Cited by
-
Use of urea and glycine betaine to quantify coupled folding and probe the burial of DNA phosphates in lac repressor-lac operator binding.Biochemistry. 2005 Dec 27;44(51):16896-911. doi: 10.1021/bi0515218. Biochemistry. 2005. PMID: 16363803 Free PMC article.
-
Isolation, sequence identification, and tissue expression profile of 3 novel porcine genes: NCF2, BCKDHB and BCKDHA.J Appl Genet. 2009;50(1):47-50. doi: 10.1007/BF03195651. J Appl Genet. 2009. PMID: 19193982
-
Folding of the protein domain hbSBD.Biophys J. 2005 Nov;89(5):3353-61. doi: 10.1529/biophysj.105.065151. Epub 2005 Aug 26. Biophys J. 2005. PMID: 16126825 Free PMC article.
References
-
- Agashe, V.R. and Udgaonkar, J.B. 1995. Thermodynamics of denaturation of barstar: Evidence for cold denaturation and evaluation of the interaction with guanidine hydrochloride. Biochemistry 34: 3286–3289. - PubMed
-
- Agashe, V.R., Schmid, F.X., and Udgaonkar, J.B. 1997. Thermodynamics of the complex protein unfolding reaction of barstar. Biochemistry 36 12288–12295. - PubMed
-
- Albert, T. and Mathews, B.W. 1987. Structure and thermal stability of phage T4 lysozyme. Methods Enzymol. 154 511–534. - PubMed
-
- Bachhawat, K., Kapoor, M., Dam, T.K., and Surolia, A. 2001. The reversible two-state unfolding of a monocot mannose-binding lectin from garlic bulbs reveals the dominant role of the dimeric interface in its stabilization. Biochemistry 40 7291–7300. - PubMed
-
- Becktel, W.J. and Schellman, J.A. 1987. Protein stability curves. Biopolymers 26 1859–1877. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

