Probing the active site of YjeE: a vital Escherichia coli protein of unknown function
- PMID: 15324301
- PMCID: PMC1134143
- DOI: 10.1042/BJ20041082
Probing the active site of YjeE: a vital Escherichia coli protein of unknown function
Abstract
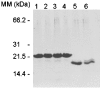
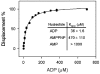
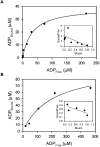
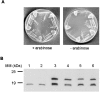
In the study described here, we have taken steps to characterize the YjeE protein, an Escherichia coli protein of unknown function that is essential for bacterial viability. YjeE represents a protein family whose members are broadly conserved in bacteria, absent from eukaryotes and contain both Walker A and B motifs, characteristic of P-loop ATPases. We have revisited the dispensability of the yjeE gene in E. coli and describe efforts to probe the function of the YjeE protein with in vitro biochemistry. We have looked critically for ATPase activity in the recombinant E. coli protein and have made vigilant use of site-directed variants in the Walker A [K41A (Lys41-->Ala) and T42A] and putative Walker B (D80Q) motifs. We noted that any hydrolysis of ATP by the wild-type E. coli protein might be attributed to background ATPase, since it was not appreciably different from that of the variants. To overcome potential contaminants, we turned to crystalline pure YjeE protein from Haemophilus influenzae that was found to hydrolyse ATP at a slow rate (kcat=1 h(-1)). We have also shown high-affinity binding to YjeE by ADP using equilibrium dialysis (K(d)=32 microM) and by fluorescence resonance energy transfer from a conserved tryptophan in YjeE to a fluorescent derivative of ADP, 2'-/3'-O-(N-methylanthraniloyl)adenosine 5'-O-diphosphate (K(d)=8 microM). Walker motif variants were notably impaired for ADP binding and T42A and D80Q mutations in yjeE were incapable of complementing the yjeE deletion strain.
Figures



References
-
- Fleischmann R. D., Adams M. D., White O., Clayton R. A., Kirkness E. F., Kerlavage A. R., Bult C. J., Tomb J. F., Dougherty B. A., Merrick J. M. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. - PubMed
-
- Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., Smith H. O., Yandell M., Evans C. A., Holt R. A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
-
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature (London) 2001;409:860–921. - PubMed
-
- Badarinarayana V., Estep P. W., 3rd, Shendure J., Edwards J., Tavazoie S., Lam F., Church G. M. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 2001;19:1060–1065. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

