Chondroitin 4-sulphotransferase-1 and chondroitin 6-sulphotransferase-1 are affected differently by uronic acid residues neighbouring the acceptor GalNAc residues
- PMID: 15324304
- PMCID: PMC1134142
- DOI: 10.1042/BJ20040965
Chondroitin 4-sulphotransferase-1 and chondroitin 6-sulphotransferase-1 are affected differently by uronic acid residues neighbouring the acceptor GalNAc residues
Abstract
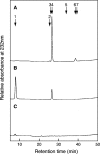
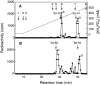
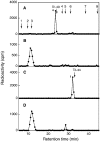
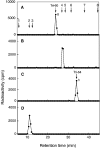
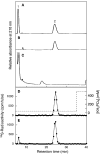
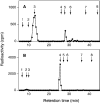
C4ST-1 (chondroitin 4-sulphotransferase-1) and C6ST-1 (chondroitin 6-sulphotransferase-1) transfer sulphate from PAPS (adenosine 3'-phosphate 5'-phosphosulphate) to positions 4 and 6 respectively of the GalNAc residues of chondroitin. We showed previously that C4ST-1 purified from rat chondrosarcoma and recombinant C4ST-1 both transfer sulphate efficiently to position 4 of the GalNAc residues of DSDS (desulphated dermatan sulphate). We report here the specificity of C4ST-1 and C6ST-1 in terms of uronic acid residue recognition around the GalNAc residue to which sulphate is transferred. When [35S]glycosaminoglycans formed from DSDS after incubation with [35S]PAPS and C4ST-1 were digested with chondroitinase ACII, a major part of the radioactivity was recovered in disaccharide fractions and the remainder distributed to tetrasaccharides and larger fractions, indicating that C4ST-1 mainly transferred sulphate to position 4 of the GalNAc residue located at the GlcA-GalNAc-GlcA sequence. Structural analysis of tetrasaccharide and larger oligosaccharide fractions indicated that C4ST-1 mainly transferred sulphate to the GalNAc residue adjacent to the reducing side of the GlcA residue. On the other hand, when [35S]glycosaminoglycans formed from DSDS after incubation with [35S]PAPS and C6ST-1 were digested with chondroitinase ACII, a major part of the radioactivity was recovered in fractions larger than hexasaccharides, indicating that C6ST-1 transferred sulphate to the GalNAc residues located in the L-iduronic acid-rich region. Structural analysis of the tetrasaccharide and larger oligosaccharide fractions indicated that C6ST-1 showed very little preference for the GalNAc residue neighbouring the GlcA residue. These results indicate that C4ST-1 and C6ST-1 differ from each other in the recognition of uronic acid residues adjacent to the targeted GalNAc residue.
Figures



Similar articles
-
A unique nonreducing terminal modification of chondroitin sulfate by N-acetylgalactosamine 4-sulfate 6-o-sulfotransferase.J Biol Chem. 2003 Oct 3;278(40):38443-52. doi: 10.1074/jbc.M306132200. Epub 2003 Jul 21. J Biol Chem. 2003. PMID: 12874280
-
Molecular cloning of squid N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase and synthesis of a unique chondroitin sulfate containing E-D hybrid tetrasaccharide structure by the recombinant enzyme.Glycobiology. 2007 Dec;17(12):1365-76. doi: 10.1093/glycob/cwm103. Epub 2007 Sep 23. Glycobiology. 2007. PMID: 17893095
-
Recognition of sulfation pattern of chondroitin sulfate by uronosyl 2-O-sulfotransferase.J Biol Chem. 2005 Nov 25;280(47):39115-23. doi: 10.1074/jbc.M508816200. Epub 2005 Sep 27. J Biol Chem. 2005. PMID: 16192264
-
Biosynthesis of chondroitin/dermatan sulfate.IUBMB Life. 2002 Oct;54(4):177-86. doi: 10.1080/15216540214923. IUBMB Life. 2002. PMID: 12512856 Review.
-
Biosynthesis and function of chondroitin sulfate.Biochim Biophys Acta. 2013 Oct;1830(10):4719-33. doi: 10.1016/j.bbagen.2013.06.006. Epub 2013 Jun 14. Biochim Biophys Acta. 2013. PMID: 23774590 Review.
Cited by
-
Inhibition of N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase by beta-D-4-O-sulfo-N-acetylgalactosaminides bearing various hydrophobic aglycons.Glycoconj J. 2010 Feb;27(2):237-48. doi: 10.1007/s10719-009-9272-7. Epub 2009 Dec 18. Glycoconj J. 2010. PMID: 20016933
-
A Review of Chondroitin Sulfate's Preparation, Properties, Functions, and Applications.Molecules. 2023 Oct 15;28(20):7093. doi: 10.3390/molecules28207093. Molecules. 2023. PMID: 37894574 Free PMC article. Review.
References
-
- Yamauchi S., Hirahara Y., Usui H., Takeda Y., Hoshino M., Fukuta M., Kimura J. H., Habuchi O. Purification and characterization of chondroitin 4-sulfotransferase from the culture medium of a rat chondrosarcoma cell line. J. Biol. Chem. 1999;274:2456–2463. - PubMed
-
- Yamauchi S., Mita S., Matsubara T., Fukuta M., Habuchi H., Kimata K., Habuchi O. Molecular cloning and expression of chondroitin 4-sulfotransferase. J. Biol. Chem. 2000;275:8975–8981. - PubMed
-
- Habuchi O., Matsui Y., Kotoya Y., Aoyama Y., Yasuda Y., Noda M. Purification of chondroitin 6-sulfotransferase secreted from cultured chick embryo chondrocytes. J. Biol. Chem. 1993;268:21968–21974. - PubMed
-
- Fukuta M., Uchimura K., Nakashima K., Kato M., Kimata K., Shinomura T., Habuchi O. Molecular cloning and expression of chick chondrocyte chondroitin 6-sulfotransferase. J. Biol. Chem. 1995;270:18575–18580. - PubMed
-
- Hiraoka N., Nakagawa H., Ong E., Akama T. O., Fukuda M. N., Fukuda M. Molecular cloning and expression of two distinct human chondroitin 4-O-sulfotransferases that belong to the HNK-1 sulfotransferase gene family. J. Biol. Chem. 2000;275:20188–20196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

