Chloroplast DNA rearrangements in Campanulaceae: phylogenetic utility of highly rearranged genomes
- PMID: 15324459
- PMCID: PMC516026
- DOI: 10.1186/1471-2148-4-27
Chloroplast DNA rearrangements in Campanulaceae: phylogenetic utility of highly rearranged genomes
Abstract
Background: The Campanulaceae (the "hare bell" or "bellflower" family) is a derived angiosperm family comprised of about 600 species treated in 35 to 55 genera. Taxonomic treatments vary widely and little phylogenetic work has been done in the family. Gene order in the chloroplast genome usually varies little among vascular plants. However, chloroplast genomes of Campanulaceae represent an exception and phylogenetic analyses solely based on chloroplast rearrangement characters support a reasonably well-resolved tree.
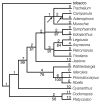
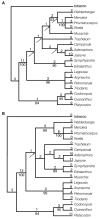
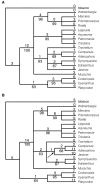
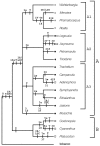
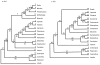
Results: Chloroplast DNA physical maps were constructed for eighteen representatives of the family. So many gene order changes have occurred among the genomes that characterizing individual mutational events was not always possible. Therefore, we examined different, novel scoring methods to prepare data matrices for cladistic analysis. These approaches yielded largely congruent results but varied in amounts of resolution and homoplasy. The strongly supported nodes were common to all gene order analyses as well as to parallel analyses based on ITS and rbcL sequence data. The results suggest some interesting and unexpected intrafamilial relationships. For example fifteen of the taxa form a derived clade; whereas the remaining three taxa--Platycodon, Codonopsis, and Cyananthus--form the basal clade. This major subdivision of the family corresponds to the distribution of pollen morphology characteristics but is not compatible with previous taxonomic treatments.
Conclusions: Our use of gene order data in the Campanulaceae provides the most highly resolved phylogeny as yet developed for a plant family using only cpDNA rearrangements. The gene order data showed markedly less homoplasy than sequence data for the same taxa but did not resolve quite as many nodes. The rearrangement characters, though relatively few in number, support robust and meaningful phylogenetic hypotheses and provide new insights into evolutionary relationships within the Campanulaceae.
Figures


References
-
- Kovanda M. Campanulaceae. In: Heywood VH, editor. In Flowering Plants of the World. New York: Mayflower Books; 1978. pp. 254–256.
-
- Takhtajan AL. Systema Magnoliophytorum. Leningrad: Nauka; 1987.
-
- de Candolle A. Monographie des Campanulêes Paris. 1830.
-
- Kolakovsky AA. System of the Campanulaceae family from the old world. Bot Zhurnal. 1987;72:1572–1579.
-
- Fedorov AA. Campanulaceae. In: Shishkin BK, editor. In Flora of the USSR. Moskva-Leningrad: Akademia Nauk SSSR; 1972. pp. 92–324.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

