Recombinant viruses and early global HIV-1 epidemic
- PMID: 15324542
- PMCID: PMC3323344
- DOI: 10.3201/eid1007.030904
Recombinant viruses and early global HIV-1 epidemic
Abstract
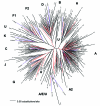
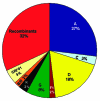
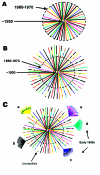
Central Africa was the epicenter of the HIV type 1 (HIV-1) pandemic. Understanding the early epidemic in the Democratic Republic of the Congo, formerly Zaire, could provide insight into how HIV evolved and assist vaccine design and intervention efforts. Using enzyme immunosorbent assays, we tested 3,988 serum samples collected in Kinshasa in the mid-1980s and confirmed seroreactivity by Western blot. Polymerase chain reaction of gag p17, env C2V3C3, and/or gp41; DNA sequencing; and genetic analyses were performed. Gene regions representing all the HIV-1 group M clades and unclassifiable sequences were found. From two or three short gene regions, 37% of the strains represented recombinant viruses, multiple infections, or both, which suggests that if whole genome sequences were available, most of these strains would have mosaic genomes. We propose that the HIV epidemic was well established in central Africa by the early 1980s and that some recombinant viruses most likely seeded the early global epidemic.
Figures
References
-
- Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, et al. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74:10498–507. 10.1128/JVI.74.22.10498-10507.2000 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
