Sizing DNA using a nanometer-diameter pore
- PMID: 15326034
- PMCID: PMC1304707
- DOI: 10.1529/biophysj.104.041814
Sizing DNA using a nanometer-diameter pore
Abstract
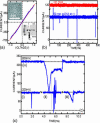
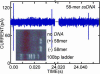
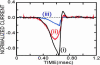
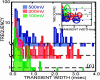
Each species from bacteria to human has a distinct genetic fingerprint. Therefore, a mechanism that detects a single molecule of DNA represents the ultimate analytical tool. As a first step in the development of such a tool, we have explored using a nanometer-diameter pore, sputtered in a nanometer-thick inorganic membrane with a tightly focused electron beam, as a transducer that detects single molecules of DNA and produces an electrical signature of the structure. When an electric field is applied across the membrane, a DNA molecule immersed in electrolyte is attracted to the pore, blocks the current through it, and eventually translocates across the membrane as verified unequivocally by gel electrophoresis. The relationship between DNA translocation and blocking current has been established through molecular dynamics simulations. By measuring the duration and magnitude of the blocking current transient, we can discriminate single-stranded from double-stranded DNA and resolve the length of the polymer.
Copyright 2004 Biophysical Society
Figures


References
-
- Allee, D. R., C. P. Umbach, and A. N. Broers. 1991. Direct nanometer scale patterning of SiO2 with electron-beam irradiation. J. Vac. Sci. Technol. B. 9:2838–2841.
-
- Bayley, H., and P. S. Cremer. 2001. Stochastic sensors inspired by biology. Nature. 413:226–230. - PubMed
-
- Bayley, H., and C. R. Martin. 2000. Resistive-pulse sensing—from microbes to molecules. Chem. Rev. 100:2575–2594. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

