A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis
- PMID: 15326298
- PMCID: PMC516574
- DOI: 10.1073/pnas.0405450101
A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis
Abstract
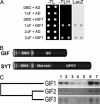
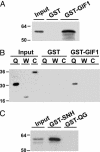
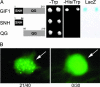
Previously, we described the AtGRF [Arabidopsis thaliana growth-regulating factor (GRF)] gene family, which encodes putative transcription factors that play a regulatory role in growth and development of leaves and cotyledons. We demonstrate here that the C-terminal region of GRF proteins has transactivation activity. In search of partner proteins for GRF1, we identified another gene family, GRF-interacting factor (GIF), which comprises three members. Sequence and molecular analysis showed that GIF1 is a functional homolog of the human SYT transcription coactivator. We found that the N-terminal region of GIF1 protein was involved in the interaction with GRF1. To understand the biological function of GIF1, we isolated a loss-of-function mutant of GIF1 and prepared transgenic plants subject to GIF1-specific RNA interference. Like grf mutants, the gif1 mutant and transgenic plants developed narrower leaves and petals than did wild-type plants, and combinations of gif1 and grf mutations showed a cooperative effect. The narrow leaf phenotype of gif1, as well as that of the grf triple mutant, was caused by a reduction in cell numbers along the leaf-width axis. We propose that GRF1 and GIF1 act as transcription activator and coactivator, respectively, and that they are part of a complex involved in regulating the growth and shape of leaves and petals.
Figures




References
-
- Kim, J. H., Choi, D. & Kende, H. (2003) Plant J. 36, 94-104. - PubMed
-
- Choi, D., Kim, J. H. & Kende, H. (2004) Plant Cell Physiol. 45, 897-904. - PubMed
-
- Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

