Genome-wide transcriptional profiling of the Escherichia coli response to a proline-rich antimicrobial peptide
- PMID: 15328082
- PMCID: PMC514742
- DOI: 10.1128/AAC.48.9.3260-3267.2004
Genome-wide transcriptional profiling of the Escherichia coli response to a proline-rich antimicrobial peptide
Abstract
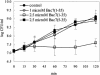
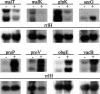
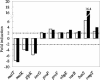
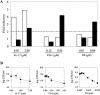
Most antimicrobial peptides (AMPs) impair the viability of target bacteria by permeabilizing bacterial membranes. However, the proline-rich AMPs have been shown to kill susceptible organisms without causing significant membrane perturbation and may act by inhibiting the activity of bacterial targets. To gain initial insight into the events that follow interaction of a proline-rich peptide with bacterial cells, we used DNA macroarray technology to monitor transcriptional alterations of Escherichia coli in response to challenge with a subinhibitory concentration of the proline-rich Bac7(1-35). Substantial changes in the expression levels of 70 bacterial genes from various functional categories were detected. Among these, 26 genes showed decreased expression, while 44 genes, including genes that are potentially involved in bacterial resistance to antimicrobials, showed increased expression. The generation of a transcriptional response under the experimental conditions used is consistent with the ability of Bac7(1-35) to interact with bacterial components and affect biological processes in this organism.
Figures

Similar articles
-
Systematic Analysis of Intracellular-targeting Antimicrobial Peptides, Bactenecin 7, Hybrid of Pleurocidin and Dermaseptin, Proline-Arginine-rich Peptide, and Lactoferricin B, by Using Escherichia coli Proteome Microarrays.Mol Cell Proteomics. 2016 Jun;15(6):1837-47. doi: 10.1074/mcp.M115.054999. Epub 2016 Feb 22. Mol Cell Proteomics. 2016. PMID: 26902206 Free PMC article.
-
The Mechanism of Killing by the Proline-Rich Peptide Bac7(1-35) against Clinical Strains of Pseudomonas aeruginosa Differs from That against Other Gram-Negative Bacteria.Antimicrob Agents Chemother. 2017 Mar 24;61(4):e01660-16. doi: 10.1128/AAC.01660-16. Print 2017 Apr. Antimicrob Agents Chemother. 2017. PMID: 28137800 Free PMC article.
-
Discovering the mechanism of action of novel antibacterial agents through transcriptional profiling of conditional mutants.Antimicrob Agents Chemother. 2005 Feb;49(2):749-59. doi: 10.1128/AAC.49.2.749-759.2005. Antimicrob Agents Chemother. 2005. PMID: 15673760 Free PMC article.
-
Proline-rich antimicrobial peptides targeting protein synthesis.Nat Prod Rep. 2017 Jul 1;34(7):702-711. doi: 10.1039/c7np00020k. Epub 2017 May 24. Nat Prod Rep. 2017. PMID: 28537612 Review.
-
The antibacterial effect of a proline-rich antibacterial peptide A3-APO.Curr Med Chem. 2009;16(30):3996-4002. doi: 10.2174/092986709789352295. Curr Med Chem. 2009. PMID: 19747127 Review.
Cited by
-
A genomic approach highlights common and diverse effects and determinants of susceptibility on the yeast Saccharomyces cerevisiae exposed to distinct antimicrobial peptides.BMC Microbiol. 2010 Nov 15;10:289. doi: 10.1186/1471-2180-10-289. BMC Microbiol. 2010. PMID: 21078184 Free PMC article.
-
Microbiological effects of sublethal levels of antibiotics.Nat Rev Microbiol. 2014 Jul;12(7):465-78. doi: 10.1038/nrmicro3270. Epub 2014 May 27. Nat Rev Microbiol. 2014. PMID: 24861036 Review.
-
Mechanisms of Action for Antimicrobial Peptides With Antibacterial and Antibiofilm Functions.Front Microbiol. 2019 Dec 12;10:2866. doi: 10.3389/fmicb.2019.02866. eCollection 2019. Front Microbiol. 2019. PMID: 31921046 Free PMC article. Review.
-
Reassessing the Host Defense Peptide Landscape.Front Chem. 2019 Feb 4;7:43. doi: 10.3389/fchem.2019.00043. eCollection 2019. Front Chem. 2019. PMID: 30778385 Free PMC article. Review.
-
Combined systems approaches reveal highly plastic responses to antimicrobial peptide challenge in Escherichia coli.PLoS Pathog. 2014 May 1;10(5):e1004104. doi: 10.1371/journal.ppat.1004104. eCollection 2014 May. PLoS Pathog. 2014. PMID: 24789011 Free PMC article.
References
-
- Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. - PubMed
-
- Bader, M. W., W. W. Navarre, W. Shiau, H. Nikaido, J. G. Frye, M. McClelland, F. C. Fang, and S. I. Miller. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219-230. - PubMed
-
- Boman, H. G. 2003. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 254:197-215. - PubMed
-
- Chan, Y. R., and R. L. Gallo. 1998. PR-39, a syndecan-inducing antimicrobial peptide, binds and affects p130(Cas). J. Biol. Chem. 273:28978-28985. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

