Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system
- PMID: 15328088
- PMCID: PMC514774
- DOI: 10.1128/AAC.48.9.3298-3304.2004
Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system
Abstract
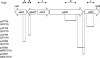
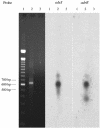
The AdeABC pump of Acinetobacter baumannii BM4454, which confers resistance to various antibiotic classes including aminoglycosides, is composed of the AdeA, AdeB, and AdeC proteins; AdeB is a member of the RND superfamily. The adeA, adeB, and adeC genes are contiguous and adjacent to adeS and adeR, which are transcribed in the opposite direction and which specify proteins homologous to sensors and regulators of two-component systems, respectively (S. Magnet, P. Courvalin, and T. Lambert, Antimicrob. Agents Chemother. 45:3375-3380, 2001). Analysis by Northern hybridization indicated that the three genes were cotranscribed, although mRNAs corresponding to adeAB and adeC were also present. Cotranscription of the two regulatory genes was demonstrated by reverse transcription-PCR. Inactivation of adeS led to aminoglycoside susceptibility. Transcripts corresponding to adeAB were not detected in susceptible A. baumannii CIP 70-10 but were present in spontaneous gentamicin-resistant mutants obtained in vitro. Analysis of these mutants revealed the substitutions Thr153-->Met in AdeS downstream from the putative His-149 site of autophosphorylation, which is presumably responsible for the loss of phosphorylase activity by the sensor, and Pro116-->Leu in AdeR at the first residue of the alpha(5) helix of the receiver domain, which is involved in interactions that control the output domain of response regulators. These mutations led to constitutive expression of the pump and, thus, to antibiotic resistance. These data indicate that the AdeABC pump is cryptic in wild A. baumannii due to stringent control by the AdeRS two-component system.
Figures


References
-
- Aiba, H., F. Nakasai, S. Mizushima, and T. Mizuno. 1989. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J. Biol. Chem. 264:14090-14094. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

