DNA ligases ensure fidelity by interrogating minor groove contacts
- PMID: 15328364
- PMCID: PMC516055
- DOI: 10.1093/nar/gkh781
DNA ligases ensure fidelity by interrogating minor groove contacts
Abstract
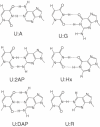
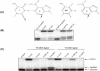
DNA ligases, found in both prokaryotes and eukaryotes, covalently link the 3'-hydroxyl and 5'-phosphate ends of duplex DNA segments. This reaction represents a completion step for DNA replication, repair and recombination. It is well established that ligases are sensitive to mispairs present on the 3' side of the ligase junction, but tolerant of mispairs on the 5' side. While such discrimination would increase the overall accuracy of DNA replication and repair, the mechanisms by which this fidelity is accomplished are as yet unknown. In this paper, we present the results of experiments with Tth ligase from Thermus thermophilus HB8 and a series of nucleoside analogs in which the mechanism of discrimination has been probed. Using a series of purine analogs substituted in the 2 and 6 positions, we establish that the apparent base pair geometry is much more important than relative base pair stability and that major groove contacts are of little importance. This result is further confirmed using 5-fluorouracil (FU) mispaired with guanine. At neutral pH, the FU:G mispair on the 3' side of a ligase junction is predominantly in a neutral wobble configuration and is poorly ligated. Increasing the solution pH increases the proportion of an ionized base pair approximating Watson-Crick geometry, substantially increasing the relative ligation efficiency. These results suggest that the ligase could distinguish Watson-Crick from mispaired geometry by probing the hydrogen bond acceptors present in the minor groove as has been proposed for DNA polymerases. The significance of minor groove hydrogen bonding interactions is confirmed with both Tth and T4 DNA ligases upon examination of base pairs containing the pyrimidine shape analog, difluorotoluene (DFT). Although DFT paired with adenine approximates Watson-Crick geometry, a minor groove hydrogen bond acceptor is lost. Consistent with this hypothesis, we observe that DFT-containing base pairs inhibit ligation when on the 3' side of the ligase junction. The NAD+-dependent ligase, Tth, is more sensitive to the DFT analog on the unligated strand whereas the ATP-dependent T4 ligase is more sensitive to substitutions in the template strand. Electrophoretic gel mobility-shift assays demonstrate that the Tth ligase binds poorly to oligonucleotide substrates containing analogs with altered minor groove contacts.
Figures





Similar articles
-
Fidelity of mispair formation and mispair extension is dependent on the interaction between the minor groove of the primer terminus and Arg668 of DNA polymerase I of Escherichia coli.Biochemistry. 2005 Apr 19;44(15):5647-59. doi: 10.1021/bi047460f. Biochemistry. 2005. PMID: 15823023
-
pH-Dependent configurations of a 5-chlorouracil-guanine base pair.Biochemistry. 2009 Dec 1;48(47):11312-8. doi: 10.1021/bi901154t. Biochemistry. 2009. PMID: 19863157 Free PMC article.
-
Minor groove recognition of the conserved G.U pair at the Tetrahymena ribozyme reaction site.Science. 1995 Feb 3;267(5198):675-9. doi: 10.1126/science.7839142. Science. 1995. PMID: 7839142
-
The steric hypothesis for DNA replication and fluorine hydrogen bonding revisited in light of structural data.Acc Chem Res. 2012 Aug 21;45(8):1237-46. doi: 10.1021/ar200303k. Epub 2012 Apr 23. Acc Chem Res. 2012. PMID: 22524491 Free PMC article. Review.
-
The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems.EMBO Rep. 2000 Jul;1(1):18-23. doi: 10.1093/embo-reports/kvd001. EMBO Rep. 2000. PMID: 11256617 Free PMC article. Review.
Cited by
-
Reconstitution of the very short patch repair pathway from Escherichia coli.J Biol Chem. 2012 Sep 21;287(39):32953-66. doi: 10.1074/jbc.M112.384321. Epub 2012 Jul 30. J Biol Chem. 2012. PMID: 22846989 Free PMC article.
-
Rational design of an XNA ligase through docking of unbound nucleic acids to toroidal proteins.Nucleic Acids Res. 2019 Jul 26;47(13):7130-7142. doi: 10.1093/nar/gkz551. Nucleic Acids Res. 2019. PMID: 31334814 Free PMC article.
-
Conformational changes of the phenyl and naphthyl isocyanate-DNA adducts during DNA replication and by minor groove binding molecules.Nucleic Acids Res. 2013 Oct;41(18):8581-90. doi: 10.1093/nar/gkt608. Epub 2013 Jul 19. Nucleic Acids Res. 2013. PMID: 23873956 Free PMC article.
-
Structure-guided mutational analysis of the OB, HhH, and BRCT domains of Escherichia coli DNA ligase.J Biol Chem. 2008 Aug 22;283(34):23343-52. doi: 10.1074/jbc.M802945200. Epub 2008 May 30. J Biol Chem. 2008. PMID: 18515356 Free PMC article.
-
Selectivity of Enzymatic Conversion of Oligonucleotide Probes during Nucleotide Polymorphism Analysis of DNA.Acta Naturae. 2010 Apr;2(1):36-53. Acta Naturae. 2010. PMID: 22649627 Free PMC article.
References
-
- Lehman I.R. (1974) DNA ligase: structure, mechanism, and function. Science, 186, 790–797. - PubMed
-
- Timson D.J., Singleton,M.R. and Wigley,D.B. (2000) DNA ligases in the repair and replication of DNA. Mutat. Res., 460, 301–318. - PubMed
-
- Tomkinson A.E. and Mackey,Z.B. (1998) Structure and function of mammalian DNA ligases. Mutat. Res., 407, 1–9. - PubMed
-
- Husain H., Tomkinson,A.E., Burkhart,W.A., Moyer,M.B., Ramos,W., Mackey,Z.B., Besterman,J.M. and Chen,J. (1995) Purification and characterization of DNA ligase III from bovine testes. Homology with DNA ligase II and vaccinia DNA ligase. J. Biol. Chem., 270, 9683–9690. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

