Redefining the structure of the mouse connexin43 gene: selective promoter usage and alternative splicing mechanisms yield transcripts with different translational efficiencies
- PMID: 15328367
- PMCID: PMC516064
- DOI: 10.1093/nar/gkh792
Redefining the structure of the mouse connexin43 gene: selective promoter usage and alternative splicing mechanisms yield transcripts with different translational efficiencies
Abstract
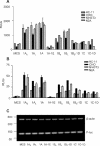
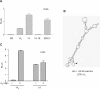
The connexin43 (cx43) gene was originally described as consisting of two exons, one coding for most of the 5'-untranslated region (5'-UTR), and the other for the protein sequence and 3'-UTR. We now report that in mouse four additional exons are expressed, all coding for novel 5'-UTRs. Altogether, we found nine different cx43 mRNA species (GenBank accession numbers NM010288, and AY427554 through AY427561) generated by differential promoter usage and alternative splicing mechanisms. The relative abundance of these different mRNAs varied with the tissue source. In addition, the different transcripts showed varying translational efficiencies in several cell lines, indicating the presence of cis-RNA elements that regulate cx43 translation. We propose that it is the promoter driving the expression of the cx43 gene that determines exon choice in the downstream splicing events in a cell-type-dependent fashion. This in turn will affect the translation efficiency of the transcript orchestrating the events that lead to the final expression profile of cx43. Since a similar organization of the cx43 gene was also observed in rat it is likely that the complex regulation of cx43 expression involving transcription, splicing and translation mechanisms is a common trait conserved during evolution.
Figures




References
-
- Bruzzone R., White,T.W. and Paul,D.L. (1996) Connections with connexins: the molecular basis of direct intercellular signaling. Eur. J. Biochem., 23, 1–27. - PubMed
-
- Bergoffen J., Scherer,S.S., Wang,S., Scott,M.O., Bone,L.J., Paul,D.L., Chen,K., Lensch,M.W., Chance,P.F. and Fischbeck,K.H. (1993) Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science, 262, 2039–2042. - PubMed
-
- Abrahms C.K., Oh,S., Ri,Y. and Bargiello,T.A. (2000) Mutations in connexin 32: the molecular and biophysical bases for the X-linked form of Charcot-Marie-Tooth disease. Brain Res. Rev., 32, 203–214. - PubMed
-
- Severs N.J. (2002) Gap junction remodeling in heart failure. J. Card. Fail., 8 (Suppl. 6), S293–S299. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions

