Recognition and accommodation at the androgen receptor coactivator binding interface
- PMID: 15328534
- PMCID: PMC509409
- DOI: 10.1371/journal.pbio.0020274
Recognition and accommodation at the androgen receptor coactivator binding interface
Abstract
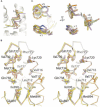
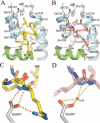
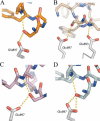
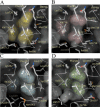
Prostate cancer is a leading killer of men in the industrialized world. Underlying this disease is the aberrant action of the androgen receptor (AR). AR is distinguished from other nuclear receptors in that after hormone binding, it preferentially responds to a specialized set of coactivators bearing aromatic-rich motifs, while responding poorly to coactivators bearing the leucine-rich "NR box" motifs favored by other nuclear receptors. Under normal conditions, interactions with these AR-specific coactivators through aromatic-rich motifs underlie targeted gene transcription. However, during prostate cancer, abnormal association with such coactivators, as well as with coactivators containing canonical leucine-rich motifs, promotes disease progression. To understand the paradox of this unusual selectivity, we have derived a complete set of peptide motifs that interact with AR using phage display. Binding affinities were measured for a selected set of these peptides and their interactions with AR determined by X-ray crystallography. Structures of AR in complex with FxxLF, LxxLL, FxxLW, WxxLF, WxxVW, FxxFF, and FxxYF motifs reveal a changing surface of the AR coactivator binding interface that permits accommodation of both AR-specific aromatic-rich motifs and canonical leucine-rich motifs. Induced fit provides perfect mating of the motifs representing the known family of AR coactivators and suggests a framework for the design of AR coactivator antagonists.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures





References
-
- Asada S, Choi Y, Uesugi M. A gene-expression inhibitor that targets an alpha-helix-mediated protein interaction. J Am Chem Soc. 2003;125:4992–4993. - PubMed
-
- Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

