Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR
- PMID: 1533056
- PMCID: PMC1904348
- DOI: 10.1126/science.1533056
Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR
Abstract
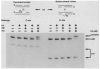
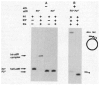
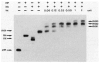
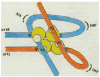
The excisive recombination reaction of bacteriophage lambda involves a specific and efficient juxtaposition of two distant higher order protein-DNA complexes on the chromosome of Escherichia coli. These complexes, which mediate synapsis and strand exchange, consist of two DNA sequences, attL and attR, the bivalent DNA binding protein Int, and the sequence-specific DNA bending proteins, IHF, Xis, and Fis. The protein-protein and protein-DNA interactions within, and between, these complexes were studied by various biochemical techniques and the patterns of synergism among pairs of mutants with marginally impaired recombination function were analyzed. The DNA bending proteins facilitated long-range tethering of high- and low-affinity DNA sites by the bivalent Int protein, and a specific map is proposed for the resulting Int bridges. These structural motifs provide a basis for postulating the mechanism of site-specific recombination and may also be relevant to other pathways in which two distant chromosomal sites become associated.
Figures
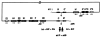
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

