Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction
- PMID: 15331581
- PMCID: PMC2291693
Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction
Abstract
Polymerase chain reaction (PCR)-based assays can target either DNA (the genome) or RNA (the transcriptome). Targeting the genome generates robust data that are informative and, most importantly, generally applicable. This is because the information contained within the genome is context-independent; i.e., generally, every normal cell contains the same DNA sequence--the same mutations and polymorphisms. The transcriptome, on the other hand, is context-dependent; i.e., the mRNA complement and level varies with physiology, pathology, or development. This makes the information contained within the transcriptome intrinsically flexible and variable. If this variability is combined with the technical limitations inherent in any reverse-transcription (RT)-PCR assay, it can be difficult to achieve not just a technically accurate but a biologically relevant result. Template quality, operator variability, the RT step itself, and subjectivity in data analysis and reporting are just a few technical aspects that make real-time RT-PCR appear to be a fragile assay that makes accurate data interpretation difficult. There can be little doubt that in the future, transcriptome-based analysis will become a routine technique. However, for the time being it remains a research tool, and it is important to recognize the considerable pitfalls associated with transcriptome analysis, with the successful application of RTPCR depending on careful experimental design, application, and validation.
Copyright 2004 ABRF
Figures

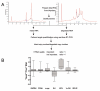
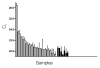


References
-
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 2000;25:169. - PubMed
-
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J Mol Endocrinol 2002;29:23–39. - PubMed
-
- Gabert J, Beillard E, van der Velden VH, et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—A Europe Against Cancer program. Leukemia 2003;17:2318–2357. - PubMed
-
- Yeung AT, Holloway BP, Adams PS, Shipley GL. Evaluation of dual-labeled fluorescent DNA probe purity versus performance in real-time PCR. Biotechniques 2004;36:266–270, 272, 274–265. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
