Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus
- PMID: 15331696
- PMCID: PMC514987
- DOI: 10.1128/JVI.78.18.9633-9644.2004
Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus
Abstract
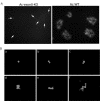
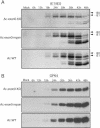
exon0 (orf141) of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is a highly conserved baculovirus gene that codes for a predicted 261-amino-acid protein. Located in the C-terminal region of EXON0 are a predicted leucine-rich coiled-coil domain and a RING finger motif. The 5' 114 nucleotides of exon0 form part of ie0, which is a spliced gene expressed at very early times postinfection, but transcriptional analysis revealed that exon0 is transcribed as a late gene. To determine the role of exon0 in the baculovirus life cycle, we used AcMNPV bacmids and generated exon0 knockout viruses (Ac-exon0-KO) by recombination in Escherichia coli. Ac-exon0-KO progressed through the very late phases in Sf9 cells, as evidenced by the development of occlusion bodies in the nuclei of the transfected or infected cells. However, production of budded virus (BV) in Ac-exon0-KO-infected cells was reduced at least 3 orders of magnitude in comparison to that in wild-type virus infection. Microscopy revealed that Ac-exon0-KO was restricted primarily to the cells initially infected, exhibiting a single-cell infection phenotype. Slot blot assays and Western blot analysis indicated that exon0 deletion did not affect the onset or levels of viral DNA replication or the expression of IE1, IE0, and GP64 prior to BV release. These results demonstrate that exon0 is required for efficient production of BV in the AcMNPV life cycle but does not affect late occlusion-derived virus.
Copyright 2004 American Society for Microbiology
Figures







References
-
- Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. - PubMed
-
- Blissard, G. W. 1996. Baculovirus-insect cell interactions. Cytotechnology 20:73-93. - PubMed
-
- Blissard, G. W., and G. F. Rohrmann. 1989. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 170:537-555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

