Discrimination among rhinovirus serotypes for a variant ICAM-1 receptor molecule
- PMID: 15331736
- PMCID: PMC514980
- DOI: 10.1128/JVI.78.18.10034-10044.2004
Discrimination among rhinovirus serotypes for a variant ICAM-1 receptor molecule
Abstract
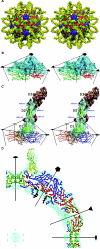
Intercellular adhesion molecule 1 (ICAM-1) is the cellular receptor for the major group of human rhinovirus serotypes, including human rhinovirus 14 (HRV14) and HRV16. A naturally occurring variant of ICAM-1, ICAM-1Kilifi, has altered binding characteristics with respect to different HRV serotypes. HRV14 binds to ICAM-1 only transiently at physiological temperatures but forms a stable complex with ICAM-1Kilifi. Conversely, HRV16 forms a stable complex with ICAM-1 but does not bind to ICAM-1Kilifi. The three-dimensional structures of HRV14 and HRV16, complexed with ICAM-1, and the structure of HRV14, complexed with ICAM-1Kilifi, have been determined by cryoelectron microscopy (cryoEM) image reconstruction to a resolution of approximately 10 angstroms. Structures determined by X-ray crystallography of both viruses and of ICAM-1 were fitted into the cryoEM density maps. The interfaces between the viruses and receptors contain extensive ionic networks. However, the interactions between the viruses and ICAM-1Kilifi contain one less salt bridge than between the viruses and ICAM-1. As HRV16 has fewer overall interactions with ICAM-1 than HRV14, the absence of this charge interaction has a greater impact on the binding of ICAM-1Kilifi to HRV16 than to HRV14.
Copyright 2004 American Society for Microbiology
Figures





Similar articles
-
Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor.EMBO J. 1999 Nov 15;18(22):6249-59. doi: 10.1093/emboj/18.22.6249. EMBO J. 1999. PMID: 10562537 Free PMC article.
-
The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand.Proc Natl Acad Sci U S A. 1998 Apr 14;95(8):4140-5. doi: 10.1073/pnas.95.8.4140. Proc Natl Acad Sci U S A. 1998. PMID: 9539703 Free PMC article.
-
Human rhinovirus 3 at 3.0 A resolution.Structure. 1996 Oct 15;4(10):1205-20. doi: 10.1016/s0969-2126(96)00128-1. Structure. 1996. PMID: 8939746
-
Review: rhinoviruses and their ICAM receptors.J Struct Biol. 1999 Dec 1;128(1):69-74. doi: 10.1006/jsbi.1999.4143. J Struct Biol. 1999. PMID: 10600561 Review.
-
Viral cell recognition and entry.Protein Sci. 1994 Oct;3(10):1712-25. doi: 10.1002/pro.5560031010. Protein Sci. 1994. PMID: 7849588 Free PMC article. Review.
Cited by
-
ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating.Proc Natl Acad Sci U S A. 2021 May 11;118(19):e2024251118. doi: 10.1073/pnas.2024251118. Proc Natl Acad Sci U S A. 2021. PMID: 33947819 Free PMC article.
-
Engineering of single Ig superfamily domain of intercellular adhesion molecule 1 (ICAM-1) for native fold and function.J Biol Chem. 2010 May 21;285(21):15906-15. doi: 10.1074/jbc.M110.104349. Epub 2010 Mar 19. J Biol Chem. 2010. PMID: 20304924 Free PMC article.
-
An analysis of the binding characteristics of a panel of recently selected ICAM-1 binding Plasmodium falciparum patient isolates.PLoS One. 2014 Oct 31;9(10):e111518. doi: 10.1371/journal.pone.0111518. eCollection 2014. PLoS One. 2014. PMID: 25360558 Free PMC article.
-
Understanding the Association of Human Rhinovirus with Asthma.Clin Vaccine Immunol. 2015 Sep 16;23(1):6-10. doi: 10.1128/CVI.00414-15. Print 2016 Jan. Clin Vaccine Immunol. 2015. PMID: 26376925 Free PMC article. Review.
-
Fighting the Common Cold: ORMDL3 in the Crosshairs?Am J Respir Cell Mol Biol. 2020 Jun;62(6):676-677. doi: 10.1165/rcmb.2020-0052ED. Am J Respir Cell Mol Biol. 2020. PMID: 32109135 Free PMC article. No abstract available.
References
-
- Arnold, E., and M. G. Rossmann. 1990. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 Å. J. Mol. Biol. 211:763-801. - PubMed
-
- Badger, J., I. Minor, M. J. Kremer, M. A. Oliveira, T. J. Smith, J. P. Griffith, D. M. A. Guerin, S. Krishnaswamy, M. Luo, M. G. Rossmann, M. A. McKinlay, G. D. Diana, F. J. Dutko, M. Fancher, R. R. Rueckert, and B. A. Heinz. 1988. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc. Natl. Acad. Sci. USA 85:3304-3308. - PMC - PubMed
-
- Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

