During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription
- PMID: 15331750
- PMCID: PMC514973
- DOI: 10.1128/JVI.78.18.10178-10186.2004
During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription
Abstract
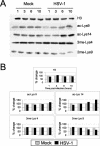
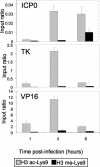
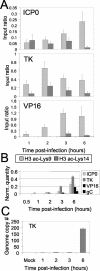
Herpes simplex virus type 1 (HSV-1) is a large (150-kb) double-stranded DNA virus that forms latent infections in neuronal cells of the human peripheral nervous system. Previous work determined that the HSV-1 genome is found in an ordered nucleosomal structure during latent infection. However, during lytic infection, it was unclear whether viral DNA was in a chromatin state. We examined HSV-1 during lytic infection using micrococcal nuclease digestion and chromatin immunoprecipitation. The HSV-1 genome is at least partially nucleosomal, although apparently not in a regular repeating structure. Analysis of histones associated with HSV-1, within both the promoter and the transcribed regions, revealed covalent amino tail modifications similar to those associated with active host mammalian genes. Certain of the modifications were detected in the temporal order expected of the immediate-early, early, and late gene classes. These data suggest that productive infection may be accompanied by acquisition of a permissive chromatin state.
Copyright 2004 American Society for Microbiology
Figures



References
-
- Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. - PubMed
-
- Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. - PubMed
-
- Boekhoudt, G. H., Z. Guo, G. W. Beresford, and J. M. Boss. 2003. Communication between NF-kappa B and Sp1 controls histone acetylation within the proximal promoter of the monocyte chemoattractant protein 1 gene. J. Immunol. 170:4139-4147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

