Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly
- PMID: 15332113
- PMCID: PMC1299141
- DOI: 10.1038/sj.embor.7400240
Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly
Abstract
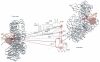
The cricket paralysis virus intergenic region internal ribosomal entry site (CrPV IGR IRES) can assemble translation initiation complexes by binding to 40S subunits without Met-tRNA(Met)(i) and initiation factors (eIFs) and then by joining directly with 60S subunits, yielding elongation-competent 80S ribosomes. Here, we report that eIF1, eIF1A and eIF3 do not significantly influence IRES/40S subunit binding but strongly inhibit subunit joining and the first elongation cycle. The IRES can avoid their inhibitory effect by its ability to bind directly to 80S ribosomes. The IRES's ability to bind to 40S subunits simultaneously with eIF1 allowed us to use directed hydroxyl radical cleavage to map its position relative to the known position of eIF1. A connecting loop in the IRES's pseudoknot (PK) III domain, part of PK II and the entire domain containing PK I are solvent-exposed and occupy the E site and regions of the P site that are usually occupied by Met-tRNA(Met)(i).
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

