Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast
- PMID: 15337848
- PMCID: PMC1370648
- DOI: 10.1261/rna.7270204
Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast
Abstract
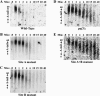
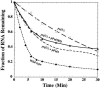
The Puf family of RNA-binding proteins regulates mRNA translation and decay via interactions with 3' untranslated regions (3' UTRs) of target mRNAs. In yeast, Puf3p binds the 3' UTR of COX17 mRNA and promotes rapid deadenylation and decay. We have investigated the sequences required for Puf3p recruitment to this 3' UTR and have identified two separate binding sites. These sites are specific for Puf3p, as they cannot bind another Puf protein, Puf5p. Both sites use a conserved UGUANAUA sequence, whereas one site contains additional sequences that enhance binding affinity. In vivo, presence of either site partially stimulates COX17 mRNA decay, but full decay regulation requires the presence of both sites. No other sequences outside the 3' UTR are required to mediate this decay regulation. The Puf repeat domain of Puf3p is sufficient not only for in vitro binding to the 3' UTR, but also in vivo stimulation of COX17 mRNA decay. These experiments indicate that the essential residues involved in mRNA decay regulation are wholly contained within this RNA-binding domain.
Copyright 2004 RNA Society
Figures





References
-
- Asaoka-Taguchi, M., Yamada, M., Nakamura, A., Hanyu, K., and Kobayashi, S. 1999. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1: 431–437. - PubMed
-
- Beers, J., Glerum, D.M., and Tzagoloff, A. 1997. Purification, characterization and localization of yeast COX17p, a mitochondrial copper shuttle. J. Biol. Chem. 272: 33191–33196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
