Neighbor-constrained segmentation with level set based 3-D deformable models
- PMID: 15338728
- PMCID: PMC2838450
- DOI: 10.1109/TMI.2004.830802
Neighbor-constrained segmentation with level set based 3-D deformable models
Abstract
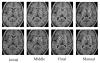
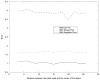
A novel method for the segmentation of multiple objects from three-dimensional (3-D) medical images using interobject constraints is presented. Our method is motivated by the observation that neighboring structures have consistent locations and shapes that provide configurations and context that aid in segmentation. We define a maximum a posteriori (MAP) estimation framework using the constraining information provided by neighboring objects to segment several objects simultaneously. We introduce a representation for the joint density function of the neighbor objects, and define joint probability distributions over the variations of the neighboring shape and position relationships of a set of training images. In order to estimate the MAP shapes of the objects, we formulate the model in terms of level set functions, and compute the associated Euler-Lagrange equations. The contours evolve both according to the neighbor prior information and the image gray level information. This method is useful in situations where there is limited interobject information as opposed to robust global atlases. In addition, we compare our level set representation of the object shape to the point distribution model. Results and validation from experiments on synthetic data and medical imagery in two-dimensional and 3-D are demonstrated.
Figures



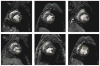
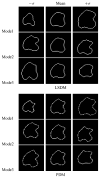
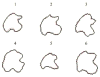
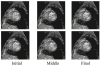




References
-
- Kass M, Witkin A, Terzopoulos D. Snakes: Active contour models. Int J Comput Vis. 1987;1:321–331.
-
- Osher S, Sethian JA. Fronts propagating with curvature-dependent speed: Algorithms based on Hamilton-Jacobi formulation. J Comp Phy. 1988;79:12–49.
-
- Chan T, Vese L. Active contours without edges. IEEE Trans Image Processing. 2001 Feb;10:266–277. - PubMed
-
- Cootes TF, Hill A, Taylor CJ, Haslam J. Use of active shape models for locating structures in medical images. Image Vis Computing. 1994 July;12(6):355–365.
-
- Staib L, Duncan J. Boundary finding with parametrically deformable models. IEEE Trans Pattern Anal Machine Intell. 1992 Nov;14:1061–1075.

