Analysis of side-chain rotamers in transmembrane proteins
- PMID: 15339811
- PMCID: PMC1304812
- DOI: 10.1529/biophysj.104.044024
Analysis of side-chain rotamers in transmembrane proteins
Abstract
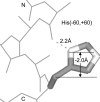
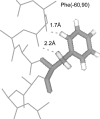
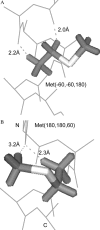
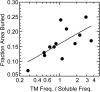
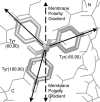
We measured the frequency of side-chain rotamers in 14 alpha-helical and 16 beta-barrel membrane protein structures and found that the membrane environment considerably perturbs the rotamer frequencies compared to soluble proteins. Although there are limited experimental data, we found statistically significant changes in rotamer preferences depending on the residue environment. Rotamer distributions were influenced by whether the residues were lipid or protein facing, and whether the residues were found near the N- or C-terminus. Hydrogen-bonding interactions with the helical backbone perturbs the rotamer populations of Ser and His. Trp and Tyr favor side-chain conformations that allow their side chains to extend their polar atoms out of the membrane core, thereby aligning the side-chain polarity gradient with the polarity gradient of the membrane. Our results demonstrate how the membrane environment influences protein structures, providing information that will be useful in the structure prediction and design of transmembrane proteins.
Figures


References
-
- Arkin, I. T., and A. T. Brunger. 1998. Statistical analysis of predicted transmembrane alpha-helices. Biochim. Biophys. Acta. 1429:113–128. - PubMed
-
- Bowie, J. U., R. Luthy, and D. Eisenberg. 1991. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 253:164–170. - PubMed
-
- Burley, S. K., and G. A. Petsko. 1988. Weakly polar interactions in proteins. Adv. Protein Chem. 39:125–189. - PubMed
-
- Chakrabarti, P., and S. Chakrabarti. 1998. C–H…O hydrogen bond involving proline residues in alpha-helices. J. Mol. Biol. 284:867–873. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

