The small nucle(ol)ar RNA cap trimethyltransferase is required for ribosome synthesis and intact nucleolar morphology
- PMID: 15340060
- PMCID: PMC515057
- DOI: 10.1128/MCB.24.18.7976-7986.2004
The small nucle(ol)ar RNA cap trimethyltransferase is required for ribosome synthesis and intact nucleolar morphology
Abstract
Nucleolar morphogenesis is a poorly defined process. Here we report that the Saccharomyces cerevisiae nucleolar trimethyl guanosine synthase I (Tgs1p), which specifically selects the m(7)G cap structure of snRNAs and snoRNAs for m(2,2,7)G conversion, is required not only for efficient pre-mRNA splicing but also for pre-rRNA processing and small ribosomal subunit synthesis. Mutational analysis indicates that the requirement for Tgs1p in pre-mRNA splicing, but not its involvement in ribosome synthesis, is dependent upon its function in cap trimethylation. In addition, we report that cells lacking Tgs1p showed a striking and unexpected loss of nucleolar structural organization. Tgs1p is not a core component of the snoRNP proteins; however, in vitro, the protein interacts with the KKD/E domain present at the carboxyl-terminal ends of several snoRNP proteins. Strains expressing versions of the snoRNPs lacking the KKD/E domain were also defective for nucleolar morphology and showed a loss of nucleolar compaction. We propose that the transient and functional interactions of Tgs1p with the abundant snoRNPs, through presumed interactions with the KKD/E domain of the snoRNP proteins, contribute substantially to the coalescence of nucleolar components. This conclusion is compatible with a model of self-organization for nucleolar assembly.
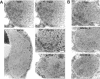
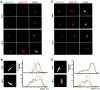
Figures




Similar articles
-
Deletion of Swm2p selectively impairs trimethylation of snRNAs by trimethylguanosine synthase (Tgs1p).FEBS Lett. 2010 Aug 4;584(15):3299-304. doi: 10.1016/j.febslet.2010.07.001. Epub 2010 Jul 15. FEBS Lett. 2010. PMID: 20621096
-
Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus.Mol Cell. 2002 Apr;9(4):891-901. doi: 10.1016/s1097-2765(02)00484-7. Mol Cell. 2002. PMID: 11983179
-
Esf2p, a U3-associated factor required for small-subunit processome assembly and compaction.Mol Cell Biol. 2005 Jul;25(13):5523-34. doi: 10.1128/MCB.25.13.5523-5534.2005. Mol Cell Biol. 2005. PMID: 15964808 Free PMC article.
-
Nucleolar RNPs: from genes to functional snoRNAs in plants.Biochem Soc Trans. 2010 Apr;38(2):672-6. doi: 10.1042/BST0380672. Biochem Soc Trans. 2010. PMID: 20298241 Review.
-
Ribosomal RNA processing and ribosome biogenesis in eukaryotes.IUBMB Life. 2004 Aug;56(8):457-65. doi: 10.1080/15216540400010867. IUBMB Life. 2004. PMID: 15545225 Review.
Cited by
-
Assembly and trafficking of box C/D and H/ACA snoRNPs.RNA Biol. 2017 Jun 3;14(6):680-692. doi: 10.1080/15476286.2016.1243646. Epub 2016 Oct 7. RNA Biol. 2017. PMID: 27715451 Free PMC article. Review.
-
General, rapid, and transcription-dependent fragmentation of nucleolar antigens in S. cerevisiae mRNA export mutants.RNA. 2008 Apr;14(4):706-16. doi: 10.1261/rna.718708. Epub 2008 Feb 7. RNA. 2008. PMID: 18258809 Free PMC article.
-
PnTgs1-like expression during reproductive development supports a role for RNA methyltransferases in the aposporous pathway.BMC Plant Biol. 2014 Nov 18;14:297. doi: 10.1186/s12870-014-0297-0. BMC Plant Biol. 2014. PMID: 25404464 Free PMC article.
-
A yeast phenomic model for the influence of Warburg metabolism on genetic buffering of doxorubicin.Cancer Metab. 2019 Oct 23;7:9. doi: 10.1186/s40170-019-0201-3. eCollection 2019. Cancer Metab. 2019. PMID: 31660150 Free PMC article.
-
CRM1 controls the composition of nucleoplasmic pre-snoRNA complexes to licence them for nucleolar transport.EMBO J. 2011 Jun 1;30(11):2205-18. doi: 10.1038/emboj.2011.128. Epub 2011 Apr 26. EMBO J. 2011. PMID: 21522132 Free PMC article.
References
-
- Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
