Regulation of human p53 activity and cell localization by alternative splicing
- PMID: 15340061
- PMCID: PMC515058
- DOI: 10.1128/MCB.24.18.7987-7997.2004
Regulation of human p53 activity and cell localization by alternative splicing
Abstract
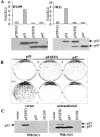
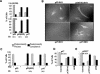
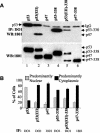
The development of cancer is a multistep process involving mutations in proto-oncogenes, tumor suppressor genes, and other genes which control cell proliferation, telomere stability, angiogenesis, and other complex traits. Despite this complexity, the cellular pathways controlled by the p53 tumor suppressor protein are compromised in most, if not all, cancers. In normal cells, p53 controls cell proliferation, senescence, and/or mediates apoptosis in response to stress, cell damage, or ectopic oncogene expression, properties which make p53 the prototype tumor suppressor gene. Defining the mechanisms of regulation of p53 activity in normal and tumor cells has therefore been a major priority in cell biology and cancer research. The present study reveals a novel and potent mechanism of p53 regulation originating through alternative splicing of the human p53 gene resulting in the expression of a novel p53 mRNA. This novel p53 mRNA encodes an N-terminally deleted isoform of p53 termed p47. As demonstrated within, p47 was able to effectively suppress p53-mediated transcriptional activity and impair p53-mediated growth suppression. It was possible to select for p53-null cells expressing p47 alone or coexpressing p53 in the presence of p47 but not cells expressing p53 alone. This showed that p47 itself does not suppress cell viability but could control p53-mediated growth suppression. Interestingly, p47 was monoubiquitinated in an Mdm2-independent manner, and this was associated with its export out of the nucleus. In the presence of p47, there was a reduction in Mdm2-mediated polyubiquitination and degradation of p53, and this was also associated with increased monoubiquitination and nuclear export of p53. The expression of p47 through alternative splicing of the p53 gene thus has a major influence over p53 activity at least in part through controlling p53 ubiquitination and cell localization.
Figures


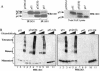
Similar articles
-
Mono- versus polyubiquitination: differential control of p53 fate by Mdm2.Science. 2003 Dec 12;302(5652):1972-5. doi: 10.1126/science.1091362. Science. 2003. PMID: 14671306
-
ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor.Cell. 2005 Jul 1;121(7):1071-83. doi: 10.1016/j.cell.2005.03.037. Cell. 2005. PMID: 15989956
-
Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4.Cancer Res. 2006 Oct 1;66(19):9502-8. doi: 10.1158/0008-5472.CAN-05-4271. Cancer Res. 2006. PMID: 17018606
-
Alternative and aberrant splicing of MDM2 mRNA in human cancer.Cancer Cell. 2002 Jul;2(1):9-15. doi: 10.1016/s1535-6108(02)00091-0. Cancer Cell. 2002. PMID: 12150820 Review.
-
Mdm2: A regulator of cell growth and death.Adv Cancer Res. 2003;89:1-34. doi: 10.1016/s0065-230x(03)01001-7. Adv Cancer Res. 2003. PMID: 14587869 Review.
Cited by
-
The Role of the p53 Protein in Stem-Cell Biology and Epigenetic Regulation.Cold Spring Harb Perspect Med. 2016 Sep 1;6(9):a026153. doi: 10.1101/cshperspect.a026153. Cold Spring Harb Perspect Med. 2016. PMID: 27352800 Free PMC article. Review.
-
p53 Isoforms in Cellular Senescence- and Ageing-Associated Biological and Physiological Functions.Int J Mol Sci. 2019 Nov 29;20(23):6023. doi: 10.3390/ijms20236023. Int J Mol Sci. 2019. PMID: 31795382 Free PMC article. Review.
-
The Influence of Quadruplex Structure in Proximity to P53 Target Sequences on the Transactivation Potential of P53 Alpha Isoforms.Int J Mol Sci. 2019 Dec 24;21(1):127. doi: 10.3390/ijms21010127. Int J Mol Sci. 2019. PMID: 31878115 Free PMC article.
-
Discovery and characterization of a novel splice variant of the p53 tumor suppressor gene in a human T cell leukemia cellline.Int J Clin Exp Pathol. 2020 May 1;13(5):1121-1135. eCollection 2020. Int J Clin Exp Pathol. 2020. PMID: 32509087 Free PMC article.
-
p53/p73 Protein Network in Colorectal Cancer and Other Human Malignancies.Cancers (Basel). 2021 Jun 9;13(12):2885. doi: 10.3390/cancers13122885. Cancers (Basel). 2021. PMID: 34207603 Free PMC article. Review.
References
-
- Boyd, S., Tsai, K., and Jacks, T. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2:563-568. - PubMed
-
- Courtois, S., G. Verhaegh, S. North, M. G. Luciani, P. Lassus, U. Hibner, M. Oren, and P. Hainaut. 2002. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counters growth suppression by wild-type p53. Oncogene 21:6722-6728. - PubMed
-
- Donehower, L. 1996. The p53-deficient mouse: a model for basic and applied cancer studies. Semin. Cancer Biol. 7:269-278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
