Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex
- PMID: 15340069
- PMCID: PMC515033
- DOI: 10.1128/MCB.24.18.8069-8079.2004
Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex
Abstract
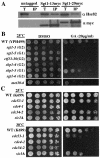
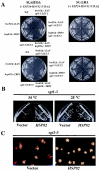
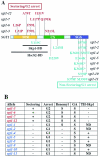
The kinetochore, which consists of DNA sequence elements and structural proteins, is essential for high-fidelity chromosome transmission during cell division. In budding yeast, Sgt1, together with Skp1, is required for assembly of the core kinetochore complex (CBF3) via Ctf13 activation. Formation of the active Ctf13-Skp1 complex also requires Hsp90, a molecular chaperone. We have found that Sgt1 interacts with Hsp90 in yeast. We also have determined that Skp1 and Hsc82 (a yeast Hsp90 protein) bind to the N-terminal region of Sgt1 that contains tetratricopeptide repeat motifs. Results of sequence and phenotypic analyses of sgt1 mutants strongly suggest that the N-terminal region containing the Hsc82-binding and Skp1-binding domains of Sgt1 is important for the kinetochore function of Sgt1. We found that Hsp90's binding to Sgt1 stimulates the binding of Sgt1 to Skp1 and that Sgt1 and Hsp90 stimulate the binding of Skp1 to Ctf13, the F-box core kinetochore protein. Our results strongly suggest that Sgt1 and Hsp90 function in assembling CBF3 by activating Skp1 and Ctf13.
Figures







References
-
- Austin, M. J., P. Muskett, K. Kahn, B. J. Feys, J. D. Jones, and J. E. Parker. 2002. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295:2077-2080. - PubMed
-
- Azevedo, C., A. Sadanandom, K. Kitagawa, A. Freialdenhoven, K. Shirasu, and P. Schulze-Lefert. 2002. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295:2073-2076. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
