Conformational changes of the flavivirus E glycoprotein
- PMID: 15341726
- PMCID: PMC4152830
- DOI: 10.1016/j.str.2004.06.019
Conformational changes of the flavivirus E glycoprotein
Abstract
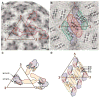
Dengue virus, a member of the Flaviviridae family, has a surface composed of 180 copies each of the envelope (E) glycoprotein and the membrane (M) protein. The crystal structure of an N-terminal fragment of E has been determined and compared with a previously described structure. The primary difference between these structures is a 10 degrees rotation about a hinge relating the fusion domain DII to domains DI and DIII. These two rigid body components were used for independent fitting of E into the cryo-electron microscopy maps of both immature and mature dengue viruses. The fitted E structures in these two particles showed a difference of 27 degrees between the two components. Comparison of the E structure in its postfusion state with that in the immature and mature virions shows a rotation approximately around the same hinge. Flexibility of E is apparently a functional requirement for assembly and infection of flaviviruses.
Figures

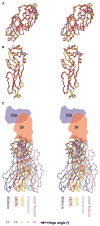


References
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

