Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation
- PMID: 15342492
- PMCID: PMC515292
- DOI: 10.1101/gad.302904
Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation
Abstract
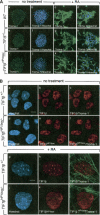
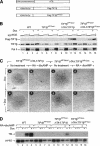
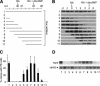
The transcriptional intermediary factor 1beta (TIF1beta) is a corepressor for KRAB-domain-containing zinc finger proteins and is believed to play essential roles in cell physiology by regulating chromatin organization at specific loci through association with chromatin remodeling and histone-modifying activities and recruitment of heterochromatin protein 1 (HP1) proteins. In this study, we have engineered a modified embryonal carcinoma F9 cell line (TIF1beta(HP1box/-)) expressing a mutated TIF1beta protein (TIF1beta(HP1box)) unable to interact with HP1 proteins. Phenotypic analysis of TIF1beta(HP1box/-) and TIF1beta(+/-) cells shows that TIF1beta-HP1 interaction is not required for differentiation of F9 cells into primitive endoderm-like (PrE) cells on retinoic acid (RA) treatment but is essential for further differentiation into parietal endoderm-like (PE) cells on addition of cAMP and for differentiation into visceral endoderm-like cells on treatment of vesicles with RA. Complementation experiments reveal that TIF1beta-HP1 interaction is essential only during a short window of time within early differentiating PrE cells to establish a selective transmittable competence to terminally differentiate on further cAMP inducing signal. Moreover, the expression of three endoderm-specific genes, GATA6, HNF4, and Dab2, is down-regulated in TIF1beta(HP1box/-) cells compared with wild-type cells during PrE differentiation. Collectively, these data demonstrate that the interaction between TIF1beta and HP1 proteins is essential for progression through differentiation by regulating the expression of endoderm differentiation master players.
Figures




References
-
- Aasland R., Gibson, T.J., and Stewart, A.F. 1995. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20: 56-59. - PubMed
-
- Ansel K.M., Lee, D.U., and Rao, A. 2003. An epigenetic view of helper T cell differentiation. Nat. Immunol. 4: 616-623. - PubMed
-
- Ayyanathan K., Lechner, M.S., Bell, P., Maul, G.G., Schultz, D.C., Yamada, Y., Tanaka, K., Torigoe, K., and Rauscher III, F.J. 2003. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes & Dev. 17: 1855-1869. - PMC - PubMed
-
- Ballas N., Battaglioli, E., Atouf, F., Andres, M.E., Chenoweth, J., Anderson, M.E., Burger, C., Moniwa, M., Davie, J.R., Bowers, W.J., et al. 2001. Regulation of neuronal traits by a novel transcriptional complex. Neuron 31: 353-365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
