Vilse, a conserved Rac/Cdc42 GAP mediating Robo repulsion in tracheal cells and axons
- PMID: 15342493
- PMCID: PMC515293
- DOI: 10.1101/gad.310204
Vilse, a conserved Rac/Cdc42 GAP mediating Robo repulsion in tracheal cells and axons
Abstract
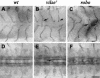
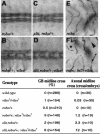
Slit proteins steer the migration of many cell types through their binding to Robo receptors, but how Robo controls cell motility is not clear. We describe the functional analysis of vilse, a Drosophila gene required for Robo repulsion in epithelial cells and axons. Vilse defines a conserved family of RhoGAPs (Rho GTPase-activating proteins), with representatives in flies and vertebrates. The phenotypes of vilse mutants resemble the tracheal and axonal phenotypes of Slit and Robo mutants at the CNS midline. Dosage-sensitive genetic interactions between vilse, slit, and robo mutants suggest that vilse is a component of robo signaling. Moreover, overexpression of Vilse in the trachea of robo mutants ameliorates the phenotypes of robo, indicating that Vilse acts downstream of Robo to mediate midline repulsion. Vilse and its human homolog bind directly to the intracellular domains of the corresponding Robo receptors and promote the hydrolysis of RacGTP and, less efficiently, of Cdc42GTP. These results together with genetic interaction experiments with robo, vilse, and rac mutants suggest a mechanism whereby Robo repulsion is mediated by the localized inactivation of Rac through Vilse.
Figures




References
-
- Adams M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. - PubMed
-
- Arber S., Barbayannis, F.A., Hanser, H., Schneider, C., Stanyon, C.A., Bernard, O., and Caroni, P. 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393: 805-809. - PubMed
-
- Bashaw G.J. and Goodman, C.S. 1999. Chimeric axon guidance receptors: The cytoplasmic domains of Slit and Netrin receptors specify attraction versus repulsion. Cell 97: 917-926. - PubMed
-
- Bashaw G.J., Kidd, T., Murray, D., Pawson, T., and Goodman, C.S. 2000. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the Roundabout receptor. Cell 101: 703-715. - PubMed
-
- Brand A.H. and Perrimon, N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401-415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
