Comparative analysis of apicomplexa and genomic diversity in eukaryotes
- PMID: 15342554
- PMCID: PMC515313
- DOI: 10.1101/gr.2615304
Comparative analysis of apicomplexa and genomic diversity in eukaryotes
Abstract
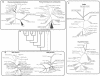
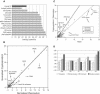
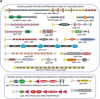
The apicomplexans Plasmodium and Cryptosporidium have developed distinctive adaptations via lineage-specific gene loss and gene innovation in the process of diverging from a common parasitic ancestor. The two lineages have acquired distinct but overlapping sets of surface protein adhesion domains typical of animal proteins, but in no case do they share multidomain architectures identical to animals. Cryptosporidium, but not Plasmodium, possesses an animal-type O-linked glycosylation pathway, along with >30 predicted surface proteins having mucin-like segments. The two parasites have notable qualitative differences in conserved protein architectures associated with chromatin dynamics and transcription. Cryptosporidium shows considerable reduction in the number of introns and a concomitant loss of spliceosomal machinery components. We also describe additional molecular characteristics distinguishing Apicomplexa from other eukaryotes for which complete genome sequences are available.
Figures


References
-
- Abrahamsen, M.S., Templeton, T.J., Enomoto, S., Abrahante, J.E., Zhu, G., Lancto, C.A., Deng, M., Liu, C., Widmer, G., Tzipori, Z., et al. 2004. The complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304: 441-445. - PubMed
-
- Aravind, L. and Subramanian, G. 1999. Origin of multicellular eukaryotes—Insights from proteome comparisons. Curr. Opin. Genet. Dev. 9: 688-694. - PubMed
WEB SITE REFERENCES
-
- http://134.84.110.219/cgi-bin/gbrowse/crypto909; C. parvum genome sequence information and annotation.
-
- http://www.cryptodb.org; C. parvum genome sequence information and annotation.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
