Recurrent sites for new centromere seeding
- PMID: 15342555
- PMCID: PMC515314
- DOI: 10.1101/gr.2608804
Recurrent sites for new centromere seeding
Abstract
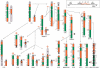
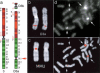
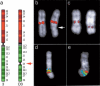
Using comparative FISH and genomics, we have studied and compared the evolution of chromosome 3 in primates and two human neocentromere cases on the long arm of this chromosome. Our results show that one of the human neocentromere cases maps to the same 3q26 chromosomal region where a new centromere emerged in a common ancestor of the Old World monkeys approximately 25-40 million years ago. Similarly, the locus in which a new centromere was seeded in the great apes' ancestor was orthologous to the site in which a new centromere emerged in the New World monkeys' ancestor. These data suggest the recurrent use of longstanding latent centromeres and that there is an inherent potential of these regions to form centromeres. The second human neocentromere case (3q24) revealed unprecedented features. The neocentromere emergence was not accompanied by any chromosomal rearrangement that usually triggers these events. Instead, it involved the functional inactivation of the normal centromere, and was present in an otherwise phenotypically normal individual who transmitted this unusual chromosome to the next generation. We propose that the formation of neocentromeres in humans and the emergence of new centromeres during the course of evolution share a common mechanism.
Figures

References
-
- Alonso, A., Mahmood, R., Li, S., Cheung, F., Yoda, K., and Warburton, P.E. 2003. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 12: 2711-2721. - PubMed
-
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. - PubMed
WEB SITE REFERENCES
-
- http://www.cs.ucsd.edu/groups/bioinformatics/GRIMM/; GRIMM software home page.
-
- www://humanparalogy.gene.cwru.edu; Segmental Duplication database.
-
- http://www.chori.org/bacpac/; Pieter de Jong libraries home page.
-
- http://www.ncbi.nlm.nih.gov/HomoloGene/; HomoloGene database.
-
- http://genome.ucsc.edu; University California Santa Cruz, Human Genome Browser Gateway.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
