DegU-P represses expression of the motility fla-che operon in Bacillus subtilis
- PMID: 15342569
- PMCID: PMC515139
- DOI: 10.1128/JB.186.18.6003-6014.2004
DegU-P represses expression of the motility fla-che operon in Bacillus subtilis
Abstract
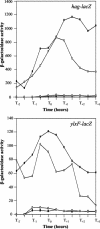
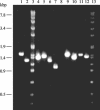
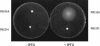
Bacillus subtilis implements several adaptive strategies to cope with nutrient limitation experienced at the end of exponential growth. The DegS-DegU two-component system is part of the network involved in the regulation of postexponential responses, such as competence development, the production of exoenzymes, and motility. The degU32(Hy) mutation extends the half-life of the phosphorylated form of DegU (DegU-P); this in turn increases the production of alkaline protease, levan-sucrase, and other exoenzymes and inhibits motility and the production of flagella. The expression of the flagellum-specific sigma factor SigD, of the flagellin gene hag, and of the fla-che operon is strongly reduced in a degU32(Hy) genetic background. To investigate the mechanism of action of DegU-P on motility, we isolated mutants of degU32(Hy) that completely suppressed the motility deficiency. The mutations were genetically mapped and characterized by PCR and sequencing. Most of the mutations were found to delete a transcriptional termination signal upstream of the main flagellar operon, fla-che, thus allowing transcriptional readthrough from the cod operon. Two additional mutations improved the sigmaA-dependent promoter sequence of the fla-che operon. Using an electrophoretic mobility shift assay, we have demonstrated that purified DegU binds specifically to the PA promoter region of the fla-che operon. The data suggest that DegU represses transcription of the fla-che operon, and they indicate a central role of the operon in regulating the synthesis and assembly of flagella.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

