Novel xylose dehydrogenase in the halophilic archaeon Haloarcula marismortui
- PMID: 15342590
- PMCID: PMC515137
- DOI: 10.1128/JB.186.18.6198-6207.2004
Novel xylose dehydrogenase in the halophilic archaeon Haloarcula marismortui
Abstract
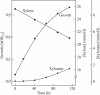
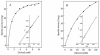
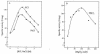
During growth of the halophilic archaeon Haloarcula marismortui on D-xylose, a specific D-xylose dehydrogenase was induced. The enzyme was purified to homogeneity. It constitutes a homotetramer of about 175 kDa and catalyzed the oxidation of xylose with both NADP+ and NAD+ as cosubstrates with 10-fold higher affinity for NADP+. In addition to D-xylose, D-ribose was oxidized at similar kinetic constants, whereas D-glucose was used with about 70-fold lower catalytic efficiency (kcat/Km). With the N-terminal amino acid sequence of the subunit, an open reading frame (ORF)-coding for a 39.9-kDA protein-was identified in the partially sequenced genome of H. marismortui. The function of the ORF as the gene designated xdh and coding for xylose dehydrogenase was proven by its functional overexpression in Escherichia coli. The recombinant enzyme was reactivated from inclusion bodies following solubilization in urea and refolding in the presence of salts, reduced and oxidized glutathione, and substrates. Xylose dehydrogenase showed the highest sequence similarity to glucose-fructose oxidoreductase from Zymomonas mobilis and other putative bacterial and archaeal oxidoreductases. Activities of xylose isomerase and xylulose kinase, the initial reactions of xylose catabolism of most bacteria, could not be detected in xylose-grown cells of H. marismortui, and the genes that encode them, xylA and xylB, were not found in the genome of H. marismortui. Thus, we propose that this first characterized archaeal xylose dehydrogenase catalyzes the initial step in xylose degradation by H. marismortui.
Figures

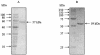


References
-
- Aoki, S., S. Ishikura, Y. Asada, N. Usami, and A. Hara. 2001. Identity of dimeric dihydrodiol dehydrogenase as NADP+-dependent d-xylose dehydrogenase in pig liver. Chemico-Biol. Interact. 130-132:775-784. - PubMed
-
- Biesterveld, S., M. D. Kok, C. Dijkema, A. J. B. Zehnder, and A. J. M. Stams. 1994. d-Xylose catabolism in Bacteriodes xylanolyticus X5-1. Arch. Microbiol. 161:521-527. - PubMed
-
- Bode, C., H. Goebell, and E. Stahler. 1968. Elimination of errors caused by turbidity in the determination of protein by the biuret method. Z. Klin. Chem. Klin. Biochem. 6:418-422. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

