Identification of a novel rat hepatic gene induced early by insulin, independently of glucose
- PMID: 15344907
- PMCID: PMC1134684
- DOI: 10.1042/BJ20040586
Identification of a novel rat hepatic gene induced early by insulin, independently of glucose
Abstract
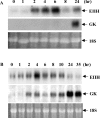
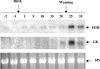
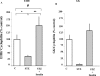
We used mRNA differential display to identify new genes induced early after exposure to insulin. Our screening strategy was based on the comparison of gene expression during the time course of insulin induction in the liver of 12-day-old suckling rats both in vivo and in vitro. A novel, early induced transcript, EIIH, was identified that encodes a 353-amino-acid protein with several features suggesting that it may be secreted or bound to membranes. EIIH is also distantly related to a variety of LRR (leucine-rich repeat) proteins. Insulin treatment increased EIIH mRNA levels in the hepatocytes of suckling, fasted adult and STZ (streptozotocin)-treated diabetic rats, where insulin was required to maintain the basal level of EIIH expression. EIIH expression was induced during the suckling/weaning transition, and remained detectable thereafter. Tissue distribution analysis in adult rats revealed a pattern of expression mainly in the liver, intestine and islets of Langerhans, closely following that of the Glut2 (glucose transporter 2), suggesting that it may play a role in carbohydrate metabolism. EIIH may be a primary target of the transcriptional regulation by insulin, and may therefore constitute a new model to study the mechanisms by which insulin acts on gene transcription.
Figures



References
-
- O'Brien R. M., Streeper R. S., Ayala J. E., Stadelmaier B. T., Hornbuckle L. A. Insulin-regulated gene expression. Biochem. Soc. Trans. 2001;29:552–558. - PubMed
-
- Fukuda H., Noguchi T., Iritani N. Transcriptional regulation of fatty acid synthase gene and ATP citrate-lyase gene by Sp1 and Sp3 in rat hepatocytes (1) FEBS Lett. 1999;464:113–117. - PubMed
-
- Nouspikel T., Iynedjian P. B. Insulin signalling and regulation of glucokinase gene expression in cultured hepatocytes. Eur. J. Biochem. 1992;210:365–373. - PubMed
-
- Tobin K. A., Ulven S. M., Schuster G. U., Steineger H. H., Andresen S. M., Gustafsson J.-Å., Nebb H. I. Liver X receptors as insulin-mediating factors in fatty acid and cholesterol biosynthesis. J. Biol. Chem. 2002;277:10691–10697. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

