Performance evaluation of commercial short-oligonucleotide microarrays and the impact of noise in making cross-platform correlations
- PMID: 15345031
- PMCID: PMC517929
- DOI: 10.1186/1471-2164-5-61
Performance evaluation of commercial short-oligonucleotide microarrays and the impact of noise in making cross-platform correlations
Abstract
Background: Despite the widespread use of microarrays, much ambiguity regarding data analysis, interpretation and correlation of the different technologies exists. There is a considerable amount of interest in correlating results obtained between different microarray platforms. To date, only a few cross-platform evaluations have been published and unfortunately, no guidelines have been established on the best methods of making such correlations. To address this issue we conducted a thorough evaluation of two commercial microarray platforms to determine an appropriate methodology for making cross-platform correlations.
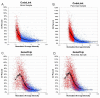
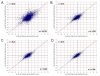
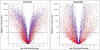
Results: In this study, expression measurements for 10,763 genes uniquely represented on Affymetrix U133A/B GeneChips and Amersham CodeLink UniSet Human 20 K microarrays were compared. For each microarray platform, five technical replicates, derived from the same total RNA samples, were labeled, hybridized, and quantified according to each manufacturers' standard protocols. The correlation coefficient (r) of differential expression ratios for the entire set of 10,763 overlapping genes was 0.62 between platforms. However, the correlation improved significantly (r = 0.79) when genes within noise were excluded. In addition to levels of inter-platform correlation, we evaluated precision, statistical-significance profiles, power, and noise levels for each microarray platform. Accuracy of differential expression was measured against real-time PCR for 25 genes and both platforms correlated well with r values of 0.92 and 0.79 for CodeLink and GeneChip, respectively.
Conclusions: As a result of this study, we recommend using only genes called 'present' in cross-platform correlations. However, as in this study, a large number of genes may be lost from the correlation due to differing levels of noise between platforms. This is an important consideration given the apparent difference in sensitivity of the two platforms. Data from microarray analysis need to be interpreted cautiously and therefore, we provide guidelines for making cross-platform correlations. In all, this study represents the most comprehensive and specifically designed comparison of short-oligonucleotide microarray platforms to date using the largest set of overlapping genes.
Figures




References
-
- Brazma A, Hingamp P, Quackenbush J, Sherlcok G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FCP, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genetics. 2001;29:365–371. doi: 10.1038/ng1201-365. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

