Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura
- PMID: 15345045
- PMCID: PMC522868
- DOI: 10.1186/gb-2004-5-9-r61
Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura
Abstract
Background: The identification of sequences that control transcription in metazoans is a major goal of genome analysis. In a previous study, we demonstrated that searching for clusters of predicted transcription factor binding sites could discover active regulatory sequences, and identified 37 regions of the Drosophila melanogaster genome with high densities of predicted binding sites for five transcription factors involved in anterior-posterior embryonic patterning. Nine of these clusters overlapped known enhancers. Here, we report the results of in vivo functional analysis of 27 remaining clusters.
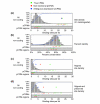
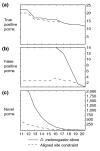
Results: We generated transgenic flies carrying each cluster attached to a basal promoter and reporter gene, and assayed embryos for reporter gene expression. Six clusters are enhancers of adjacent genes: giant, fushi tarazu, odd-skipped, nubbin, squeeze and pdm2; three drive expression in patterns unrelated to those of neighboring genes; the remaining 18 do not appear to have enhancer activity. We used the Drosophila pseudoobscura genome to compare patterns of evolution in and around the 15 positive and 18 false-positive predictions. Although conservation of primary sequence cannot distinguish true from false positives, conservation of binding-site clustering accurately discriminates functional binding-site clusters from those with no function. We incorporated conservation of binding-site clustering into a new genome-wide enhancer screen, and predict several hundred new regulatory sequences, including 85 adjacent to genes with embryonic patterns.
Conclusions: Measuring conservation of sequence features closely linked to function--such as binding-site clusterin--makes better use of comparative sequence data than commonly used methods that examine only sequence identity.
Figures




References
-
- Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

