Global nucleosome occupancy in yeast
- PMID: 15345046
- PMCID: PMC522869
- DOI: 10.1186/gb-2004-5-9-r62
Global nucleosome occupancy in yeast
Abstract
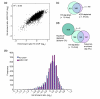
Background: Although eukaryotic genomes are generally thought to be entirely chromatin-associated, the activated PHO5 promoter in yeast is largely devoid of nucleosomes. We systematically evaluated nucleosome occupancy in yeast promoters by immunoprecipitating nucleosomal DNA and quantifying enrichment by microarrays.
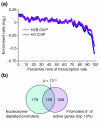
Results: Nucleosome depletion is observed in promoters that regulate active genes and/or contain multiple evolutionarily conserved motifs that recruit transcription factors. The Rap1 consensus was the only binding motif identified in a completely unbiased search of nucleosome-depleted promoters. Nucleosome depletion in the vicinity of Rap1 consensus sites in ribosomal protein gene promoters was also observed by real-time PCR and micrococcal nuclease digestion. Nucleosome occupancy in these regions was increased by the small molecule rapamycin or, in the case of the RPS11B promoter, by removing the Rap1 consensus sites.
Conclusions: The presence of transcription factor-binding motifs is an important determinant of nucleosome depletion. Most motifs are associated with marked depletion only when they appear in combination, consistent with a model in which transcription factors act collaboratively to exclude nucleosomes and gain access to target sites in the DNA. In contrast, Rap1-binding sites cause marked depletion under steady-state conditions. We speculate that nucleosome depletion enables Rap1 to define chromatin domains and alter them in response to environmental cues.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

