An Ambystoma mexicanum EST sequencing project: analysis of 17,352 expressed sequence tags from embryonic and regenerating blastema cDNA libraries
- PMID: 15345051
- PMCID: PMC522874
- DOI: 10.1186/gb-2004-5-9-r67
An Ambystoma mexicanum EST sequencing project: analysis of 17,352 expressed sequence tags from embryonic and regenerating blastema cDNA libraries
Abstract
Background: The ambystomatid salamander, Ambystoma mexicanum (axolotl), is an important model organism in evolutionary and regeneration research but relatively little sequence information has so far been available. This is a major limitation for molecular studies on caudate development, regeneration and evolution. To address this lack of sequence information we have generated an expressed sequence tag (EST) database for A. mexicanum.
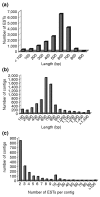
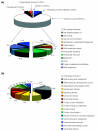
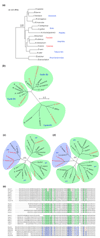
Results: Two cDNA libraries, one made from stage 18-22 embryos and the other from day-6 regenerating tail blastemas, generated 17,352 sequences. From the sequenced ESTs, 6,377 contigs were assembled that probably represent 25% of the expressed genes in this organism. Sequence comparison revealed significant homology to entries in the NCBI non-redundant database. Further examination of this gene set revealed the presence of genes involved in important cell and developmental processes, including cell proliferation, cell differentiation and cell-cell communication. On the basis of these data, we have performed phylogenetic analysis of key cell-cycle regulators. Interestingly, while cell-cycle proteins such as the cyclin B family display expected evolutionary relationships, the cyclin-dependent kinase inhibitor 1 gene family shows an unusual evolutionary behavior among the amphibians.
Conclusions: Our analysis reveals the importance of a comprehensive sequence set from a representative of the Caudata and illustrates that the EST sequence database is a rich source of molecular, developmental and regeneration studies. To aid in data mining, the ESTs have been organized into an easily searchable database that is freely available online.
Figures


References
-
- Shubin NWD. Phylogeny, variation, and morphological Integration. Am Zool. 1996;36:51–60.
-
- Roth G, Nishikawa KC, Wake DB. Genome size, secondary simplification, and the evolution of the brain in salamanders. Brain Behav Evol. 1997;50:50–59. - PubMed
-
- Animal Genome Size Database http://www.genomesize.com
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

