Phenotypic and molecular typing of Salmonella strains reveals different contamination sources in two commercial pig slaughterhouses
- PMID: 15345414
- PMCID: PMC520922
- DOI: 10.1128/AEM.70.9.5305-5314.2004
Phenotypic and molecular typing of Salmonella strains reveals different contamination sources in two commercial pig slaughterhouses
Abstract
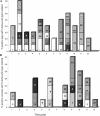
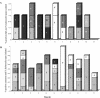
This study aimed to define the origin of Salmonella contamination on swine carcasses and the distribution of Salmonella serotypes in two commercial slaughterhouses during normal activity. Salmonellae were isolated from carcasses, from colons and mesenteric lymph nodes of individual pigs, and from the slaughterhouse environment. All strains were serotyped; Salmonella enterica serotype Typhimurium and Salmonella enterica serotype Derby isolates were additionally typed beyond the serotype level by pulsed-field gel electrophoresis (PFGE) and antibiotic resistance profiling (ARP); and a subset of 31 serotype Typhimurium strains were additionally phage typed. PFGE and ARP had the same discriminative possibility. Phage typing in combination with PFGE could give extra information for some strains. In one slaughterhouse, 21% of the carcasses were contaminated, reflecting a correlation with the delivery of infected pigs. Carcass contamination did not result only from infection of the corresponding pig; only 25% of the positive carcasses were contaminated with the same serotype or genotype found in the corresponding feces or mesenteric lymph nodes. In the other slaughterhouse, 70% of the carcasses were contaminated, and only in 4% was the same genotype or serotype detected as in the feces of the corresponding pigs. The other positive carcasses in both slaughterhouses were contaminated by genotypes present in the feces or lymph nodes of pigs slaughtered earlier that day or from dispersed sources in the environment. In slaughterhouses, complex contamination cycles may be present, resulting in the isolation of many different genotypes circulating in the environment due to the supply of positive animals and in the contamination of carcasses, probably through aerosols.
Figures
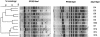

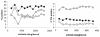
Similar articles
-
Unveiling contamination sources and dissemination routes of Salmonella sp. in pigs at a Portuguese slaughterhouse through macrorestriction profiling by pulsed-field gel electrophoresis.Int J Food Microbiol. 2006 Jul 1;110(1):77-84. doi: 10.1016/j.ijfoodmicro.2006.01.046. Epub 2006 Jul 7. Int J Food Microbiol. 2006. PMID: 16828912
-
Detection and characterization of Salmonella in lairage, on pig carcasses and intestines in five slaughterhouses.Int J Food Microbiol. 2011 Jan 31;145(1):279-86. doi: 10.1016/j.ijfoodmicro.2011.01.009. Epub 2011 Jan 12. Int J Food Microbiol. 2011. PMID: 21276632
-
Investigation of Salmonella enterica in Sardinian slaughter pigs: prevalence, serotype and genotype characterization.Int J Food Microbiol. 2011 Dec 2;151(2):201-9. doi: 10.1016/j.ijfoodmicro.2011.08.025. Epub 2011 Aug 31. Int J Food Microbiol. 2011. PMID: 21940060
-
Six-Year Follow-up of Slaughterhouse Surveillance (2008-2013): The Catalan Slaughterhouse Support Network (SESC).Vet Pathol. 2016 May;53(3):532-44. doi: 10.1177/0300985815593125. Epub 2015 Jul 13. Vet Pathol. 2016. PMID: 26169387 Review.
-
Why must we rush to bury our dead (pigs): The option of excarnation by exposure.Can Vet J. 2021 Dec;62(12):1309-1314. Can Vet J. 2021. PMID: 34857967 Free PMC article. Review.
Cited by
-
First Complete Genome Sequence of a Salmonella enterica subsp. enterica Serovar Derby Strain Associated with Pork in France.Genome Announc. 2015 Jul 30;3(4):e00853-15. doi: 10.1128/genomeA.00853-15. Genome Announc. 2015. PMID: 26227607 Free PMC article.
-
Salmonella in the pork production chain and its impact on human health in the European Union.Epidemiol Infect. 2017 Jun;145(8):1513-1526. doi: 10.1017/S095026881700036X. Epub 2017 Feb 28. Epidemiol Infect. 2017. PMID: 28241896 Free PMC article. Review.
-
Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates.J Clin Microbiol. 2006 Jul;44(7):2449-57. doi: 10.1128/JCM.00019-06. J Clin Microbiol. 2006. PMID: 16825363 Free PMC article.
-
Serotype distribution, antimicrobial susceptibility, antimicrobial resistance genes and virulence genes of Salmonella isolated from a pig slaughterhouse in Yangzhou, China.AMB Express. 2019 Dec 28;9(1):210. doi: 10.1186/s13568-019-0936-9. AMB Express. 2019. PMID: 31884559 Free PMC article.
-
Polyphyletic Nature of Salmonella enterica Serotype Derby and Lineage-Specific Host-Association Revealed by Genome-Wide Analysis.Front Microbiol. 2018 May 17;9:891. doi: 10.3389/fmicb.2018.00891. eCollection 2018. Front Microbiol. 2018. PMID: 29867804 Free PMC article.
References
-
- Baggesen, D. L., and F. M. Aarestrup. 1998. Characterisation of recently emerged multiple antibiotic-resistant Salmonella enterica serovar Typhimurium DT104 and other multiresistant phage types from Danish pig herds. Vet. Rec. 25:95-97. - PubMed
-
- Berends, B. R., F. Van Knapen, J. M. Snijders, and D. A. Mossel. 1997. Identification and quantification of risk factors regarding Salmonella spp. on pork carcasses. Int. J. Food Microbiol. 36:199-206. - PubMed
-
- Borch, E., T. Nesbakken, and H. Christensen. 1996. Hazard identification in swine slaughter with respect to foodborne bacteria. Int. J. Food Microbiol. 30:9-25. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

