Integration of microbial ecology and statistics: a test to compare gene libraries
- PMID: 15345436
- PMCID: PMC520927
- DOI: 10.1128/AEM.70.9.5485-5492.2004
Integration of microbial ecology and statistics: a test to compare gene libraries
Abstract
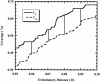
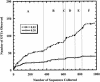
Libraries of 16S rRNA genes provide insight into the membership of microbial communities. Statistical methods help to determine whether differences in library composition are artifacts of sampling or are due to underlying differences in the communities from which they are derived. To contribute to a growing statistical framework for comparing 16S rRNA libraries, we present a computer program, integral -LIBSHUFF, which calculates the integral form of the Cramér-von Mises statistic. This implementation builds upon the LIBSHUFF program, which uses an approximation of the statistic and makes a number of modifications that improve precision and accuracy. Once integral -LIBSHUFF calculates the P values, when pairwise comparisons are tested at the 0.05 level, the probability of falsely identifying a significant P value is 0.098 for a study with two libraries, 0.265 for three libraries, and 0.460 for four libraries. The potential negative effects of making the multiple pairwise comparisons necessitate correcting for the increased likelihood that differences between treatments are due to chance and do not reflect biological differences. Using integral -LIBSHUFF, we found that previously published 16S rRNA gene libraries constructed from Scottish and Wisconsin soils contained different bacterial lineages. We also analyzed the published libraries constructed for the zebrafish gut microflora and found statistically significant changes in the community during development of the host. These analyses illustrate the power of integral -LIBSHUFF to detect differences between communities, providing the basis for ecological inference about the association of soil productivity or host gene expression and microbial community composition.
Figures

References
-
- Alain, K., M. Olagnon, D. Desbruyeres, A. Page, G. Barbier, S. K. Juniper, J. Querellou, and M. A. Cambon-Bonavita. 2002. Phylogenetic characterization of the bacterial assemblage associated with mucous secretions of the hydrothermal vent polychaete Paralvinella palmiformis. FEMS Microbiol. Ecol. 42:463-476. - PubMed
-
- Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Methodol. 57:289-300.
-
- Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

