Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones
- PMID: 15345440
- PMCID: PMC520897
- DOI: 10.1128/AEM.70.9.5522-5527.2004
Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones
Abstract
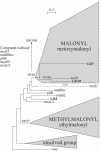
The metagenomic approach provides direct access to diverse unexplored genomes, especially from uncultivated bacteria in a given environment. This diversity can conceal many new biosynthetic pathways. Type I polyketide synthases (PKSI) are modular enzymes involved in the biosynthesis of many natural products of industrial interest. Among the PKSI domains, the ketosynthase domain (KS) was used to screen a large soil metagenomic library containing more than 100,000 clones to detect those containing PKS genes. Over 60,000 clones were screened, and 139 clones containing KS domains were detected. A 700-bp fragment of the KS domain was sequenced for 40 of 139 randomly chosen clones. None of the 40 protein sequences were identical to those found in public databases, and nucleic sequences were not redundant. Phylogenetic analyses were performed on the protein sequences of three metagenomic clones to select the clones which one can predict to produce new compounds. Two PKS-positive clones do not belong to any of the 23 published PKSI included in the analysis, encouraging further analyses on these two clones identified by the selection process.
Figures

References
-
- Aparicio, J. F., R. Fouces, M. V. Mendes, N. Olivera, and J. F. Martin. 2000. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem. Biol. 7:895-905. - PubMed
-
- Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. - PubMed
-
- Courtois, S., A. Frostegard, P. Goransson, G. Depret, P. Jeannin, and P. Simonet. 2001. Quantification of bacterial subgroups in soil: comparison of DNA extracted directly from soil or from cells previously released by density gradient centrifugation. Environ. Microbiol. 3:431-439. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

