Lactococcal plasmid pNP40 encodes a novel, temperature-sensitive restriction-modification system
- PMID: 15345443
- PMCID: PMC520859
- DOI: 10.1128/AEM.70.9.5546-5556.2004
Lactococcal plasmid pNP40 encodes a novel, temperature-sensitive restriction-modification system
Abstract
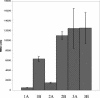
A novel restriction-modification system, designated LlaJI, was identified on pNP40, a naturally occurring 65-kb plasmid from Lactococcus lactis. The system comprises four adjacent similarly oriented genes that are predicted to encode two m(5)C methylases and two restriction endonucleases. The LlaJI system, when cloned into a low-copy-number vector, was shown to confer resistance against representatives of the three most common lactococcal phage species. This phage resistance phenotype was found to be strongly temperature dependent, being most effective at 19 degrees C. A functional analysis confirmed that the predicted methylase-encoding genes, llaJIM1 and llaJIM2, were both required to mediate complete methylation, while the assumed restriction enzymes, specified by llaJIR1 and llaJIR2, were both necessary for the complete restriction phenotype. A Northern blot analysis revealed that the four LlaJI genes are part of a 6-kb operon and that the relative abundance of the LlaJI-specific mRNA in the cells does not appear to contribute to the observed temperature-sensitive profile. This was substantiated by use of a LlaJI promoter-lacZ fusion, which further revealed that the LlaJI operon appears to be subject to transcriptional regulation by an as yet unidentified element(s) encoded by pNP40.
Figures
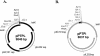



References
-
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Bourniquel, A. A., and T. A. Bickle. 2002. Complex restriction enzymes: NTP-driven molecular motors. Biochimie 84:1047-1059. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

